A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis
- PMID: 22103005
- PMCID: PMC3084211
- DOI: 10.1371/journal.pgen.1002058
A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis
Abstract
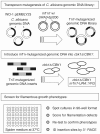
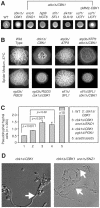
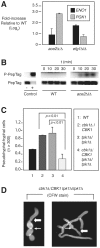
The morphogenetic transition between yeast and filamentous forms of the human fungal pathogen Candida albicans is regulated by a variety of signaling pathways. How these pathways interact to orchestrate morphogenesis, however, has not been as well characterized. To address this question and to identify genes that interact with the Regulation of Ace2 and Morphogenesis (RAM) pathway during filamentation, we report the first large-scale genetic interaction screen in C. albicans.Our strategy for this screen was based on the concept of complex haploinsufficiency (CHI). A heterozygous mutant of CBK1(cbk1Δ/CBK1), a key RAM pathway protein kinase, was subjected to transposon-mediated, insertional mutagenesis. The resulting double heterozygous mutants (6,528 independent strains) were screened for decreased filamentation on SpiderMedium (SM). From the 441 mutants showing altered filamentation, 139 transposon insertion sites were sequenced,yielding 41 unique CBK1-interacting genes. This gene set was enriched in transcriptional targets of Ace2 and, strikingly, the cAMP-dependent protein kinase A (PKA) pathway, suggesting an interaction between these two pathways. Further analysis indicates that the RAM and PKA pathways co-regulate a common set of genes during morphogenesis and that hyperactivation of the PKA pathway may compensate for loss of RAM pathway function. Our data also indicate that the PKA–regulated transcription factor Efg1 primarily localizes to yeast phase cells while the RAM–pathway regulated transcription factor Ace2 localizes to daughter nuclei of filamentous cells, suggesting that Efg1 and Ace2 regulate a common set of genes at separate stages of morphogenesis. Taken together, our observations indicate that CHI–based screening is a useful approach to genetic interaction analysis in C. albicans and support a model in which these two pathways regulate a common set of genes at different stages of filamentation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

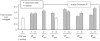


References
-
- Vazquez JA, Sobel JD. Candidiasis. In: Dismukes WE, Pappas PG, Sobel JD, editors. Clinical Mycology. Oxford: Oxford University Press; 2003. pp. 143–188.
-
- Lo HJ, Kohler JR, DiDomenico B, Loebenber D, Cacciapuoti A, et al. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. - PubMed
-
- Kumamoto CA, Vinces MD. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 2005;7:1546–1554. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

