Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA
- PMID: 22103478
- PMCID: PMC3279155
- DOI: 10.1021/ja207407n
Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA
Abstract
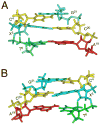
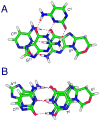
Diastereomeric 8,5'-cyclopurine 2'-deoxynucleosides, containing a covalent bond between the deoxyribose and the purine base, represent an important class of DNA damage induced by ionizing radiation. The 8,5'-cyclo-2'-deoxyguanosine lesion (cdG) has been recently reported to be a strong block of replication and highly mutagenic in Escherichia coli. The 8,5'-cyclopurine-2'-deoxyriboses are suspected to play a role in the etiology of neurodegeneration in xeroderma pigmentosum patients. These lesions cannot be repaired by base excision repair, but they are substrates for nucleotide excision repair. The structure of an oligodeoxynucleotide duplex containing a site-specific S-cdG lesion placed opposite dC in the complementary strand was obtained by molecular dynamics calculations restrained by distance and dihedral angle restraints obtained from NMR spectroscopy. The S-cdG deoxyribose exhibited the O4'-exo (west) pseudorotation. Significant perturbations were observed for the β, γ, and χ torsion angles of the S-cdG nucleoside. Watson-Crick base pairing was conserved at the S-cdG·dC pair. However, the O4'-exo pseudorotation of the S-cdG deoxyribose perturbed the helical twist and base pair stacking at the lesion site and the 5'-neighbor dC·dG base pair. Thermodynamic destabilization of the duplex measured by UV melting experiments correlated with base stacking and structural perturbations involving the modified S-cdG·dC and 3'- neighbor dT·dA base pairs. These perturbations may be responsible for both the genotoxicity of this lesion and its ability to be recognized by nucleotide excision repair.
© 2011 American Chemical Society
Figures






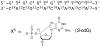
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

