Determining novel functions of Arabidopsis 14-3-3 proteins in central metabolic processes
- PMID: 22104211
- PMCID: PMC3253775
- DOI: 10.1186/1752-0509-5-192
Determining novel functions of Arabidopsis 14-3-3 proteins in central metabolic processes
Abstract
Background: 14-3-3 proteins are considered master regulators of many signal transduction cascades in eukaryotes. In plants, 14-3-3 proteins have major roles as regulators of nitrogen and carbon metabolism, conclusions based on the studies of a few specific 14-3-3 targets.
Results: In this study, extensive novel roles of 14-3-3 proteins in plant metabolism were determined through combining the parallel analyses of metabolites and enzyme activities in 14-3-3 overexpression and knockout plants with studies of protein-protein interactions. Decreases in the levels of sugars and nitrogen-containing-compounds and in the activities of known 14-3-3-interacting-enzymes were observed in 14-3-3 overexpression plants. Plants overexpressing 14-3-3 proteins also contained decreased levels of malate and citrate, which are intermediate compounds of the tricarboxylic acid (TCA) cycle. These modifications were related to the reduced activities of isocitrate dehydrogenase and malate dehydrogenase, which are key enzymes of TCA cycle. In addition, we demonstrated that 14-3-3 proteins interacted with one isocitrate dehydrogenase and two malate dehydrogenases. There were also changes in the levels of aromatic compounds and the activities of shikimate dehydrogenase, which participates in the biosynthesis of aromatic compounds.
Conclusion: Taken together, our findings indicate that 14-3-3 proteins play roles as crucial tuners of multiple primary metabolic processes including TCA cycle and the shikimate pathway.
Figures

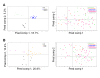

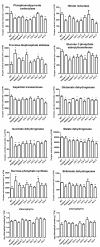


Similar articles
-
The Extra-Pathway Interactome of the TCA Cycle: Expected and Unexpected Metabolic Interactions.Plant Physiol. 2018 Jul;177(3):966-979. doi: 10.1104/pp.17.01687. Epub 2018 May 23. Plant Physiol. 2018. PMID: 29794018 Free PMC article.
-
Two mitochondrial phosphatases, PP2c63 and Sal2, are required for posttranslational regulation of the TCA cycle in Arabidopsis.Mol Plant. 2021 Jul 5;14(7):1104-1118. doi: 10.1016/j.molp.2021.03.023. Epub 2021 Mar 31. Mol Plant. 2021. PMID: 33798747
-
Tricarboxylic acid cycle enzyme activities in a mouse model of methylmalonic aciduria.Mol Genet Metab. 2019 Dec;128(4):444-451. doi: 10.1016/j.ymgme.2019.10.007. Epub 2019 Oct 17. Mol Genet Metab. 2019. PMID: 31648943 Free PMC article.
-
Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues.Plant Cell Environ. 2012 Jan;35(1):1-21. doi: 10.1111/j.1365-3040.2011.02332.x. Epub 2011 Jun 20. Plant Cell Environ. 2012. PMID: 21477125 Review.
-
The shikimate pathway: review of amino acid sequence, function and three-dimensional structures of the enzymes.Crit Rev Microbiol. 2015 Jun;41(2):172-89. doi: 10.3109/1040841X.2013.813901. Epub 2013 Aug 6. Crit Rev Microbiol. 2015. PMID: 23919299 Review.
Cited by
-
Ion Homeostasis and Metabolome Analysis of Arabidopsis 14-3-3 Quadruple Mutants to Salt Stress.Front Plant Sci. 2021 Sep 13;12:697324. doi: 10.3389/fpls.2021.697324. eCollection 2021. Front Plant Sci. 2021. PMID: 34589094 Free PMC article.
-
A Member of the 14-3-3 Gene Family in Brachypodium distachyon, BdGF14d, Confers Salt Tolerance in Transgenic Tobacco Plants.Front Plant Sci. 2017 Mar 13;8:340. doi: 10.3389/fpls.2017.00340. eCollection 2017. Front Plant Sci. 2017. PMID: 28348575 Free PMC article.
-
Cytological and proteomic analyses of horsetail (Equisetum arvense L.) spore germination.Front Plant Sci. 2015 Jun 17;6:441. doi: 10.3389/fpls.2015.00441. eCollection 2015. Front Plant Sci. 2015. PMID: 26136760 Free PMC article.
-
Genome-Wide Identification, Phylogeny, and Expression Analyses of the 14-3-3 Family Reveal Their Involvement in the Development, Ripening, and Abiotic Stress Response in Banana.Front Plant Sci. 2016 Sep 22;7:1442. doi: 10.3389/fpls.2016.01442. eCollection 2016. Front Plant Sci. 2016. PMID: 27713761 Free PMC article.
-
Protein Phosphatase (PP2C9) Induces Protein Expression Differentially to Mediate Nitrogen Utilization Efficiency in Rice under Nitrogen-Deficient Condition.Int J Mol Sci. 2018 Sep 19;19(9):2827. doi: 10.3390/ijms19092827. Int J Mol Sci. 2018. PMID: 30235789 Free PMC article.
References
-
- Cotelle V, Mackintosh C. Do 14-3-3s regulate 'resource allocation in crops'? Ann Appl Biol. 2001;138:1–7. doi: 10.1111/j.1744-7348.2001.tb00079.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

