How, when and why proteins collapse: the relation to folding
- PMID: 22104965
- PMCID: PMC3288525
- DOI: 10.1016/j.sbi.2011.10.005
How, when and why proteins collapse: the relation to folding
Abstract
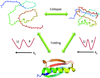
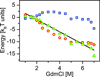
Unfolded proteins under strongly denaturing conditions are highly expanded. However, when the conditions are more close to native, an unfolded protein may collapse to a compact globular structure distinct from the folded state. This transition is akin to the coil-globule transition of homopolymers. Single-molecule FRET experiments have been particularly conducive in revealing the collapsed state under conditions of coexistence with the folded state. The collapse can be even more readily observed in natively unfolded proteins. Time-resolved studies, using FRET and small-angle scattering, have shown that the collapse transition is a very fast event, probably occurring on the submicrosecond time scale. The forces driving collapse are likely to involve both hydrophobic and backbone interactions. The loss of configurational entropy during collapse makes the unfolded state less stable compared to the folded state, thus facilitating folding.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

References
-
- de Gennes P-G. Scaling concepts in polymer physics. Cornell university press: Ithaca; 1979.
-
- Grosberg AY, Kokhlov AR. Statistical Physics of Macromolecules. AIP Press: New York; 1994.
-
- Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. - PubMed
-
- Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12491–12496. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

