Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration
- PMID: 22105172
- PMCID: PMC3226005
- DOI: 10.1172/JCI59261
Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration
Abstract
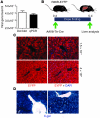
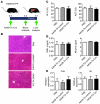
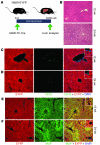
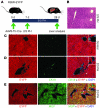
Recent evidence has contradicted the prevailing view that homeostasis and regeneration of the adult liver are mediated by self duplication of lineage-restricted hepatocytes and biliary epithelial cells. These new data suggest that liver progenitor cells do not function solely as a backup system in chronic liver injury; rather, they also produce hepatocytes after acute injury and are in fact the main source of new hepatocytes during normal hepatocyte turnover. In addition, other evidence suggests that hepatocytes are capable of lineage conversion, acting as precursors of biliary epithelial cells during biliary injury. To test these concepts, we generated a hepatocyte fate-tracing model based on timed and specific Cre recombinase expression and marker gene activation in all hepatocytes of adult Rosa26 reporter mice with an adenoassociated viral vector. We found that newly formed hepatocytes derived from preexisting hepatocytes in the normal liver and that liver progenitor cells contributed minimally to acute hepatocyte regeneration. Further, we found no evidence that biliary injury induced conversion of hepatocytes into biliary epithelial cells. These results therefore restore the previously prevailing paradigms of liver homeostasis and regeneration. In addition, our new vector system will be a valuable tool for timed, efficient, and specific loop out of floxed sequences in hepatocytes.
Figures




Comment in
-
On the origin of the liver.J Clin Invest. 2011 Dec;121(12):4630-3. doi: 10.1172/JCI59652. Epub 2011 Nov 21. J Clin Invest. 2011. PMID: 22105167 Free PMC article.
-
Phenotypic fidelity (or not?) of epithelial cells in the liver.Hepatology. 2012 Jun;55(6):2024-7. doi: 10.1002/hep.25703. Hepatology. 2012. PMID: 22407729 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

