Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway
- PMID: 22105349
- PMCID: PMC3257566
- DOI: 10.1083/jcb.201103045
Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway
Abstract
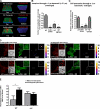
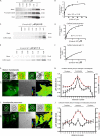
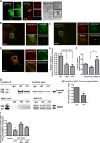
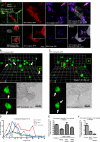
Invadopodia are invasive protrusions with proteolytic activity uniquely found in tumor cells. Cortactin phosphorylation is a key step during invadopodia maturation, regulating Nck1 binding and cofilin activity. The precise mechanism of cortactin-dependent cofilin regulation and the roles of this pathway in invadopodia maturation and cell invasion are not fully understood. We provide evidence that cortactin-cofilin binding is regulated by local pH changes at invadopodia that are mediated by the sodium-hydrogen exchanger NHE1. Furthermore, cortactin tyrosine phosphorylation mediates the recruitment of NHE1 to the invadopodium compartment, where it locally increases the pH to cause the release of cofilin from cortactin. We show that this mechanism involving cortactin phosphorylation, local pH increase, and cofilin activation regulates the dynamic cycles of invadopodium protrusion and retraction and is essential for cell invasion in 3D. Together, these findings identify a novel pH-dependent regulation of cell invasion.
Figures


References
-
- Artym V.V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K.M., Mueller S.C. 2006. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66:3034–3043 10.1158/0008-5472.CAN-05-2177 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

