Structure and function of glutamate receptor amino terminal domains
- PMID: 22106178
- PMCID: PMC3300046
- DOI: 10.1113/jphysiol.2011.213850
Structure and function of glutamate receptor amino terminal domains
Abstract
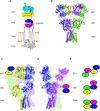
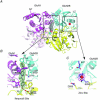
The amino terminal domain (ATD) of ionotropic glutamate receptor (iGluR) subunits resides at the extracellular region distal to the membrane. The ATD is structurally and functionally the most divergent region of the iGluR subunits. Structural studies on full-length GluA2 and the ATDs from three iGluR subfamilies have shed light on how the ATD facilitates subunit assembly, accommodates allosteric modulator compounds, and controls gating properties. Here recent developments in structural and functional studies on iGluR ATDs are reviewed.
Figures


References
-
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. - PubMed
-
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. - PubMed
-
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. - PubMed
-
- Atlason PT, Garside ML, Meddows E, Whiting P, McIlhinney RA. N-Methyl-D-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J Biol Chem. 2007;282:25299–25307. - PubMed
-
- Carron C, Jullien A, Bucher B. Synthesis and pharmacological properties of a series of 2-piperidino alkanol derivatives. Arzneimittelforschung. 1971;21:1992–1998. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

