Beta structure motifs of islet amyloid polypeptides identified through surface-mediated assemblies
- PMID: 22106265
- PMCID: PMC3241769
- DOI: 10.1073/pnas.1102971108
Beta structure motifs of islet amyloid polypeptides identified through surface-mediated assemblies
Abstract
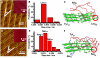
We report here the identification of the key sites for the beta structure motifs of the islet amyloid polypeptide (IAPP) analogs by using scanning tunneling microscopy (STM). Duplex folding structures in human IAPP(8-37) (hIAPP(8-37)) assembly were observed featuring a hairpin structure. The multiplicity in rIAPP assembly structures indicates the polydispersity of the rat IAPP(8-37) (rIAPP(8-37)) beta-like motifs. The bimodal length distribution of beta structure motifs for rIAPP(8-37) R18H indicates the multiple beta segments linked by turns. The IAPP(8-37) analogs share common structure motifs of IAPP(8-17) and IAPP(26-37) with the most probable key sites at positions around Ser(19)/Ser(20) and Gly(24). These observations reveal the similar amyloid formation tendency in the C and N terminus segments because of the sequence similarity, while the differences in specific amino acids at each key site manifest the effect of sequence variations. The results could be beneficial for studying structural polymorphism of amyloidal peptides with multiple beta structure motifs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Structural and energetic insight into the cross-seeding amyloid assemblies of human IAPP and rat IAPP.J Phys Chem B. 2014 Jun 26;118(25):7026-36. doi: 10.1021/jp5022246. Epub 2014 Jun 12. J Phys Chem B. 2014. PMID: 24892388
-
β-Hairpin of Islet Amyloid Polypeptide Bound to an Aggregation Inhibitor.Sci Rep. 2016 Sep 19;6:33474. doi: 10.1038/srep33474. Sci Rep. 2016. PMID: 27641459 Free PMC article.
-
Comparative molecular dynamics study of human islet amyloid polypeptide (IAPP) and rat IAPP oligomers.Biochemistry. 2013 Feb 12;52(6):1089-100. doi: 10.1021/bi301525e. Epub 2013 Jan 29. Biochemistry. 2013. PMID: 23331123
-
Islet amyloid: from fundamental biophysics to mechanisms of cytotoxicity.FEBS Lett. 2013 Apr 17;587(8):1106-18. doi: 10.1016/j.febslet.2013.01.046. Epub 2013 Feb 1. FEBS Lett. 2013. PMID: 23380070 Free PMC article. Review.
-
Islet amyloid and type 2 diabetes: from molecular misfolding to islet pathophysiology.Biochim Biophys Acta. 2001 Nov 29;1537(3):179-203. doi: 10.1016/s0925-4439(01)00078-3. Biochim Biophys Acta. 2001. PMID: 11731221 Review.
Cited by
-
Peptide conformation and oligomerization characteristics of surface-mediated assemblies revealed by molecular dynamics simulations and scanning tunneling microscopy.RSC Adv. 2019 Dec 13;9(70):41345-41350. doi: 10.1039/c9ra09320f. eCollection 2019 Dec 9. RSC Adv. 2019. PMID: 35540063 Free PMC article.
-
Impact of Electronic Polarization on Preformed, β-Strand Rich Homogenous and Heterogenous Amyloid Oligomers.J Comput Biophys Chem. 2022 Jun;21(4):449-460. doi: 10.1142/s2737416521420059. Epub 2021 Dec 29. J Comput Biophys Chem. 2022. PMID: 35756548 Free PMC article.
-
Harnessing the Activation of Toll-Like Receptor 2/6 by Self-Assembled Cross-β Fibrils to Design Adjuvanted Nanovaccines.Nanomaterials (Basel). 2020 Oct 7;10(10):1981. doi: 10.3390/nano10101981. Nanomaterials (Basel). 2020. PMID: 33036404 Free PMC article.
-
Rationally Designed Protein Building Blocks for Programmable Hierarchical Architectures.Front Chem. 2020 Oct 29;8:587975. doi: 10.3389/fchem.2020.587975. eCollection 2020. Front Chem. 2020. PMID: 33195088 Free PMC article. Review.
-
Single-molecule visualization determines conformational substate ensembles in β-sheet-rich peptide fibrils.Sci Adv. 2023 Jul 7;9(27):eadg7943. doi: 10.1126/sciadv.adg7943. Epub 2023 Jul 5. Sci Adv. 2023. PMID: 37406110 Free PMC article.
References
-
- Cooper GJS, Willis AC, Leighton B. Amylin hormone. Nature. 1989;340:272–272. - PubMed
-
- Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. - PubMed
-
- Kayed R, et al. Conformational transitions of islet amyloid poly-peptide (IAPP) in amyloid formation in vitro. J Mol Biol. 1999;287:781–796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

