Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes
- PMID: 22106369
- PMCID: PMC3290782
- DOI: 10.1101/gr.126417.111
Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes
Abstract
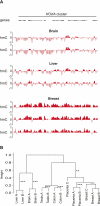
The discovery of substantial amounts of 5-hydroxymethylcytosine (5hmC), formed by the oxidation of 5-methylcytosine (5mC), in various mouse tissues and human embryonic stem (ES) cells has necessitated a reevaluation of our knowledge of 5mC/5hmC patterns and functions in mammalian cells. Here, we investigate the tissue specificity of both the global levels and locus-specific distribution of 5hmC in several human tissues and cell lines. We find that global 5hmC content of normal human tissues is highly variable, does not correlate with global 5mC content, and decreases rapidly as cells from normal tissue adapt to cell culture. Using tiling microarrays to map 5hmC levels in DNA from normal human tissues, we find that 5hmC patterns are tissue specific; unsupervised hierarchical clustering based solely on 5hmC patterns groups independent biological samples by tissue type. Moreover, in agreement with previous studies, we find 5hmC associated primarily, but not exclusively, with the body of transcribed genes, and that within these genes 5hmC levels are positively correlated with transcription levels. However, using quantitative 5hmC-qPCR, we find that the absolute levels of 5hmC for any given gene are primarily determined by tissue type, gene expression having a secondary influence on 5hmC levels. That is, a gene transcribed at a similar level in several different tissues may have vastly different levels of 5hmC (>20-fold) dependent on tissue type. Our findings highlight tissue type as a major modifier of 5hmC levels in expressed genes and emphasize the importance of using quantitative analyses in the study of 5hmC levels.
Figures




References
-
- Aflatoonian B, Ruban L, Shamsuddin S, Baker D, Andrews P, Moore H 2010. Generation of Sheffield (Shef) human embryonic stem cell lines using a microdrop culture system. In Vitro Cell Dev Biol Anim 46: 236–241 - PubMed
-
- Bird A 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 - PubMed
-
- Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42: 1093–1100 - PubMed
-
- Cleveland W 1979. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc 74: 829–836
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
