Cdc7-Dbf4 is a gene-specific regulator of meiotic transcription in yeast
- PMID: 22106412
- PMCID: PMC3255779
- DOI: 10.1128/MCB.06032-11
Cdc7-Dbf4 is a gene-specific regulator of meiotic transcription in yeast
Abstract
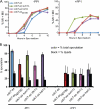
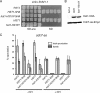
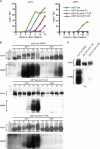
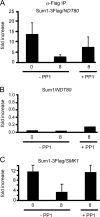
Meiosis divides the chromosome number of the cell in half by having two rounds of chromosome segregation follow a single round of chromosome duplication. The first meiotic division is unique in that homologous pairs of sister chromatids segregate to opposite poles. Recent work in budding and fission yeast has shown that the cell cycle kinase, Cdc7-Dbf4, is required for many meiosis-specific chromosomal functions necessary for proper disjunction at meiosis I. This work reveals another role for Cdc7 in meiosis as a gene-specific regulator of the global transcription factor, Ndt80, which is required for exit from pachytene and entry into the meiotic divisions in budding yeast. Cdc7-Dbf4 promotes NDT80 transcription by relieving repression mediated by a complex of Sum1, Rfm1, and a histone deacetylase, Hst1. Sum1 exhibits meiosis-specific Cdc7-dependent phosphorylation, and mass spectrometry analysis reveals a dynamic and complex pattern of phosphorylation events, including four constitutive cyclin-dependent kinase (Cdk1) sites and 11 meiosis-specific Cdc7-Dbf4-dependent sites. Analysis of various phosphorylation site mutants suggests that Cdc7 functions with both Cdk1 and the meiosis-specific kinase Ime2 to control this critical transition point during meiosis.
Figures



References
-
- Allers T, Lichten M. 2001. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106:47–57 - PubMed
-
- Bailis JM, Bernard P, Antonelli R, Allshire RC, Forsburg SL. 2003. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 5:1111–1116 - PubMed
-
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. 2006. A probability-based approach for high throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24:1285–1292 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
