Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis
- PMID: 22108385
- PMCID: PMC3225972
- DOI: 10.1128/mBio.00257-11
Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis
Abstract
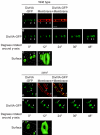
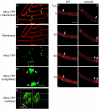
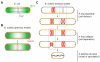
The assembly of the cell division machinery at midcell is a critical step of cytokinesis. Many rod-shaped bacteria position septa using nucleoid occlusion, which prevents division over the chromosome, and the Min system, which prevents division near the poles. Here we examined the in vivo assembly of the Bacillus subtilis MinCD targeting proteins DivIVA, a peripheral membrane protein that preferentially localizes to negatively curved membranes and resembles eukaryotic tropomyosins, and MinJ, which recruits MinCD to DivIVA. We used structured illumination microscopy to demonstrate that both DivIVA and MinJ localize as double rings that flank the septum and first appear early in septal biosynthesis. The subsequent recruitment of MinCD to these double rings would separate the Min proteins from their target, FtsZ, spatially regulating Min activity and allowing continued cell division. Curvature-based localization would also provide temporal regulation, since DivIVA and the Min proteins would localize to midcell after the onset of division. We use time-lapse microscopy and fluorescence recovery after photobleaching to demonstrate that DivIVA rings are highly stable and are constructed from newly synthesized DivIVA molecules. After cell division, DivIVA rings appear to collapse into patches at the rounded cell poles of separated cells, with little or no incorporation of newly synthesized subunits. Thus, changes in cell architecture mediate both the initial recruitment of DivIVA to sites of cell division and the subsequent collapse of these rings into patches (or rings of smaller diameter), while curvature-based localization of DivIVA spatially and temporally regulates Min activity.
Importance: The Min systems of Escherichia coli and Bacillus subtilis both inhibit FtsZ assembly, but one key difference between these two species is that whereas the E. coli Min proteins localize to the poles, the B. subtilis proteins localize to nascent division sites by interaction with DivIVA and MinJ. It is unclear how MinC activity at midcell is regulated to prevent it from interfering with FtsZ engaged in medial cell division. We used superresolution microscopy to demonstrate that DivIVA and MinJ, which localize MinCD, assemble double rings that flank active division sites and septa. This curvature-based localization mechanism holds MinCD away from the FtsZ ring at midcell, and we propose that this spatial organization is the primary mechanism by which MinC activity is regulated to allow division at midcell. Curvature-based localization also conveys temporal regulation, since it ensures that MinC localizes after the onset of division.
Figures




Similar articles
-
Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division.Genes Dev. 2008 Dec 15;22(24):3475-88. doi: 10.1101/gad.1732408. Genes Dev. 2008. PMID: 19141479 Free PMC article.
-
Septal membrane localization by C-terminal amphipathic α-helices of MinD in Bacillus subtilis mutant cells lacking MinJ or DivIVA.Genes Genet Syst. 2017 Oct 18;92(2):81-98. doi: 10.1266/ggs.16-00054. Epub 2017 Jun 30. Genes Genet Syst. 2017. PMID: 28674273
-
A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD.Mol Microbiol. 2008 Dec;70(6):1556-69. doi: 10.1111/j.1365-2958.2008.06501.x. Epub 2008 Oct 23. Mol Microbiol. 2008. PMID: 19019154
-
Division site selection in rod-shaped bacteria.Curr Opin Microbiol. 2009 Dec;12(6):683-8. doi: 10.1016/j.mib.2009.10.002. Epub 2009 Nov 1. Curr Opin Microbiol. 2009. PMID: 19884039 Review.
-
Cytokinesis in bacteria.Microbiol Mol Biol Rev. 2003 Mar;67(1):52-65, table of contents. doi: 10.1128/MMBR.67.1.52-65.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626683 Free PMC article. Review.
Cited by
-
ATP hydrolysis by a domain related to translation factor GTPases drives polymerization of a static bacterial morphogenetic protein.Proc Natl Acad Sci U S A. 2013 Jan 8;110(2):E151-60. doi: 10.1073/pnas.1210554110. Epub 2012 Dec 24. Proc Natl Acad Sci U S A. 2013. PMID: 23267091 Free PMC article.
-
The ParA/MinD family puts things in their place.Trends Microbiol. 2012 Sep;20(9):411-8. doi: 10.1016/j.tim.2012.05.002. Epub 2012 Jun 4. Trends Microbiol. 2012. PMID: 22672910 Free PMC article. Review.
-
Bactofilin-mediated organization of the ParABS chromosome segregation system in Myxococcus xanthus.Nat Commun. 2017 Nov 28;8(1):1817. doi: 10.1038/s41467-017-02015-z. Nat Commun. 2017. PMID: 29180656 Free PMC article.
-
Comparing divisome organization between vegetative and sporulating Bacillus subtilis at the nanoscale using DNA-PAINT.Sci Adv. 2024 Jan 12;10(2):eadk5847. doi: 10.1126/sciadv.adk5847. Epub 2024 Jan 10. Sci Adv. 2024. PMID: 38198550 Free PMC article.
-
Dynamics of the Bacillus subtilis Min System.mBio. 2021 Apr 13;12(2):e00296-21. doi: 10.1128/mBio.00296-21. mBio. 2021. PMID: 33849976 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
