A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis
- PMID: 22108404
- PMCID: PMC3246337
- DOI: 10.1105/tpc.111.089029
A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis
Abstract
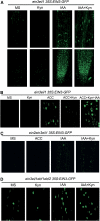
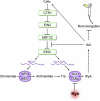
The interactions between phytohormones are crucial for plants to adapt to complex environmental changes. One example is the ethylene-regulated local auxin biosynthesis in roots, which partly contributes to ethylene-directed root development and gravitropism. Using a chemical biology approach, we identified a small molecule, l-kynurenine (Kyn), which effectively inhibited ethylene responses in Arabidopsis thaliana root tissues. Kyn application repressed nuclear accumulation of the ETHYLENE INSENSITIVE3 (EIN3) transcription factor. Moreover, Kyn application decreased ethylene-induced auxin biosynthesis in roots, and TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1/TRYPTOPHAN AMINOTRANSFERASE RELATEDs (TAA1/TARs), the key enzymes in the indole-3-pyruvic acid pathway of auxin biosynthesis, were identified as the molecular targets of Kyn. Further biochemical and phenotypic analyses revealed that Kyn, being an alternate substrate, competitively inhibits TAA1/TAR activity, and Kyn treatment mimicked the loss of TAA1/TAR functions. Molecular modeling and sequence alignments suggested that Kyn effectively and selectively binds to the substrate pocket of TAA1/TAR proteins but not those of other families of aminotransferases. To elucidate the destabilizing effect of Kyn on EIN3, we further found that auxin enhanced EIN3 nuclear accumulation in an EIN3 BINDING F-BOX PROTEIN1 (EBF1)/EBF2-dependent manner, suggesting the existence of a positive feedback loop between auxin biosynthesis and ethylene signaling. Thus, our study not only reveals a new level of interactions between ethylene and auxin pathways but also offers an efficient method to explore and exploit TAA1/TAR-dependent auxin biosynthesis.
Figures







References
-
- Abel S., Nguyen M.D., Chow W., Theologis A. (1995). ACS4, a primary indoleacetic acid-responsive gene encoding 1-aminocyclopropane-1-carboxylate synthase in Arabidopsis thaliana. Structural characterization, expression in Escherichia coli, and expression characteristics in response to auxin [corrected]. J. Biol. Chem. 270: 19093–19099 Erratum. J. Biol. Chem. 270: 26020 - PubMed
-
- Alonso J.M., Hirayama T., Roman G., Nourizadeh S., Ecker J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

