The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells
- PMID: 22108792
- PMCID: PMC3261715
- DOI: 10.1038/msb.2011.84
The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells
Abstract
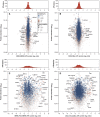
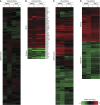
Assessing relevant molecular differences between human-induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs) is important, given that such differences may impact their potential therapeutic use. Controversy surrounds recent gene expression studies comparing hiPSCs and hESCs. Here, we present an in-depth quantitative mass spectrometry-based analysis of hESCs, two different hiPSCs and their precursor fibroblast cell lines. Our comparisons confirmed the high similarity of hESCs and hiPSCS at the proteome level as 97.8% of the proteins were found unchanged. Nevertheless, a small group of 58 proteins, mainly related to metabolism, antigen processing and cell adhesion, was found significantly differentially expressed between hiPSCs and hESCs. A comparison of the regulated proteins with previously published transcriptomic studies showed a low overlap, highlighting the emerging notion that differences between both pluripotent cell lines rather reflect experimental conditions than a recurrent molecular signature.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, Bello PA, Benvenisty N, Berry LS, Bevan S, Blum B, Brooking J, Chen KG, Choo ABH, Churchill GA, Corbel M, Damjanov I, Draper JS, Dvorak P, Emanuelsson K et al. (2007) Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25: 803–816 - PubMed
-
- Baker DEC, Harrison NJ, Maltby E, Smith K, Moore HD, Shaw PJ, Heath PR, Holden H, Andrews PW (2007) Adaptation to culture of human embryonic stem cells and oncogenesis in vivo. Nat Biotechnol 25: 207–215 - PubMed
-
- Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447: 407–412 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

