PDBe: Protein Data Bank in Europe
- PMID: 22110033
- PMCID: PMC3245096
- DOI: 10.1093/nar/gkr998
PDBe: Protein Data Bank in Europe
Abstract
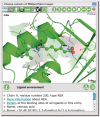
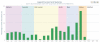
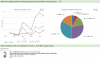
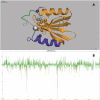
The Protein Data Bank in Europe (PDBe; pdbe.org) is a partner in the Worldwide PDB organization (wwPDB; wwpdb.org) and as such actively involved in managing the single global archive of biomacromolecular structure data, the PDB. In addition, PDBe develops tools, services and resources to make structure-related data more accessible to the biomedical community. Here we describe recently developed, extended or improved services, including an animated structure-presentation widget (PDBportfolio), a widget to graphically display the coverage of any UniProt sequence in the PDB (UniPDB), chemistry- and taxonomy-based PDB-archive browsers (PDBeXplore), and a tool for interactive visualization of NMR structures, corresponding experimental data as well as validation and analysis results (Vivaldi).
Figures


References
-
- Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. - PubMed
-
- Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr. 2008;A64:88–95. - PubMed
-
- Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

