Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells
- PMID: 22110401
- PMCID: PMC3217011
- DOI: 10.1371/journal.pbio.1001196
Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells
Abstract
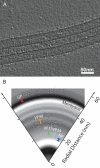
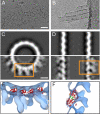
Several major human pathogens, including the filoviruses, paramyxoviruses, and rhabdoviruses, package their single-stranded RNA genomes within helical nucleocapsids, which bud through the plasma membrane of the infected cell to release enveloped virions. The virions are often heterogeneous in shape, which makes it difficult to study their structure and assembly mechanisms. We have applied cryo-electron tomography and sub-tomogram averaging methods to derive structures of Marburg virus, a highly pathogenic filovirus, both after release and during assembly within infected cells. The data demonstrate the potential of cryo-electron tomography methods to derive detailed structural information for intermediate steps in biological pathways within intact cells. We describe the location and arrangement of the viral proteins within the virion. We show that the N-terminal domain of the nucleoprotein contains the minimal assembly determinants for a helical nucleocapsid with variable number of proteins per turn. Lobes protruding from alternate interfaces between each nucleoprotein are formed by the C-terminal domain of the nucleoprotein, together with viral proteins VP24 and VP35. Each nucleoprotein packages six RNA bases. The nucleocapsid interacts in an unusual, flexible "Velcro-like" manner with the viral matrix protein VP40. Determination of the structures of assembly intermediates showed that the nucleocapsid has a defined orientation during transport and budding. Together the data show striking architectural homology between the nucleocapsid helix of rhabdoviruses and filoviruses, but unexpected, fundamental differences in the mechanisms by which the nucleocapsids are then assembled together with matrix proteins and initiate membrane envelopment to release infectious virions, suggesting that the viruses have evolved different solutions to these conserved assembly steps.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
Marburg virus structure revealed in detail.PLoS Biol. 2011 Nov;9(11):e1001198. doi: 10.1371/journal.pbio.1001198. Epub 2011 Nov 15. PLoS Biol. 2011. PMID: 22110402 Free PMC article.
References
-
- Lamb R. Mononegavirales. In: Knipe D, Howley P, editors. Fields Virology. Fifth Edition ed. Philadelphia, USA: Lippincott Williams and Wilkins; 2007. 1357
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

