Role of transcription factor modifications in the pathogenesis of insulin resistance
- PMID: 22110478
- PMCID: PMC3205681
- DOI: 10.1155/2012/716425
Role of transcription factor modifications in the pathogenesis of insulin resistance
Abstract
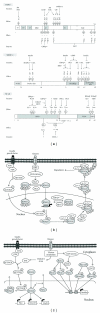
Non-alcoholic fatty liver disease (NAFLD) is characterized by fat accumulation in the liver not due to alcohol abuse. NAFLD is accompanied by variety of symptoms related to metabolic syndrome. Although the metabolic link between NAFLD and insulin resistance is not fully understood, it is clear that NAFLD is one of the main cause of insulin resistance. NAFLD is shown to affect the functions of other organs, including pancreas, adipose tissue, muscle and inflammatory systems. Currently efforts are being made to understand molecular mechanism of interrelationship between NAFLD and insulin resistance at the transcriptional level with specific focus on post-translational modification (PTM) of transcription factors. PTM of transcription factors plays a key role in controlling numerous biological events, including cellular energy metabolism, cell-cycle progression, and organ development. Cell type- and tissue-specific reversible modifications include lysine acetylation, methylation, ubiquitination, and SUMOylation. Moreover, phosphorylation and O-GlcNAcylation on serine and threonine residues have been shown to affect protein stability, subcellular distribution, DNA-binding affinity, and transcriptional activity. PTMs of transcription factors involved in insulin-sensitive tissues confer specific adaptive mechanisms in response to internal or external stimuli. Our understanding of the interplay between these modifications and their effects on transcriptional regulation is growing. Here, we summarize the diverse roles of PTMs in insulin-sensitive tissues and their involvement in the pathogenesis of insulin resistance.
Figures
Similar articles
-
Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease.Nutrients. 2013 May 10;5(5):1544-60. doi: 10.3390/nu5051544. Nutrients. 2013. PMID: 23666091 Free PMC article. Review.
-
Insulin resistance in nonalcoholic fatty liver disease.Curr Pharm Des. 2010 Jun;16(17):1941-51. doi: 10.2174/138161210791208875. Curr Pharm Des. 2010. PMID: 20370677 Review.
-
Non-alcoholic fatty liver disease, insulin resistance, metabolic syndrome and their association with vascular risk.Metabolism. 2021 Jun;119:154770. doi: 10.1016/j.metabol.2021.154770. Epub 2021 Apr 14. Metabolism. 2021. PMID: 33864798 Review.
-
Ectopic fat, insulin resistance and non-alcoholic fatty liver disease.Proc Nutr Soc. 2013 Nov;72(4):412-9. doi: 10.1017/S0029665113001249. Epub 2013 May 14. Proc Nutr Soc. 2013. PMID: 23668723 Review.
-
Nonalcoholic Fatty liver disease is associated with increased carotid intima-media thickness only in type 2 diabetic subjects with insulin resistance.J Clin Endocrinol Metab. 2014 May;99(5):1879-84. doi: 10.1210/jc.2013-4133. Epub 2014 Feb 10. J Clin Endocrinol Metab. 2014. PMID: 24512497
Cited by
-
Cardiac Development and Transcription Factors: Insulin Signalling, Insulin Resistance, and Intrauterine Nutritional Programming of Cardiovascular Disease.J Nutr Metab. 2018 Feb 1;2018:8547976. doi: 10.1155/2018/8547976. eCollection 2018. J Nutr Metab. 2018. PMID: 29484207 Free PMC article. Review.
-
Characterization of Osterix protein stability and physiological role in osteoblast differentiation.PLoS One. 2013;8(2):e56451. doi: 10.1371/journal.pone.0056451. Epub 2013 Feb 15. PLoS One. 2013. PMID: 23457570 Free PMC article.
-
Integrated analysis of transcription factors and targets co-expression profiles reveals reduced correlation between transcription factors and target genes in cancer.Funct Integr Genomics. 2019 Jan;19(1):191-204. doi: 10.1007/s10142-018-0636-6. Epub 2018 Sep 24. Funct Integr Genomics. 2019. PMID: 30251028
-
Melatonin and Melatonergic Influence on Neuronal Transcription Factors: Implications for the Development of Novel Therapies for Neurodegenerative Disorders.Curr Neuropharmacol. 2020;18(7):563-577. doi: 10.2174/1570159X18666191230114339. Curr Neuropharmacol. 2020. PMID: 31885352 Free PMC article. Review.
-
Utilizing a Comprehensive Immunoprecipitation Enrichment System to Identify an Endogenous Post-translational Modification Profile for Target Proteins.J Vis Exp. 2018 Jan 8;(131):56912. doi: 10.3791/56912. J Vis Exp. 2018. PMID: 29364248 Free PMC article.
References
-
- Darnell JE., Jr. Variety in the level of gene control in eukaryotic cells. Nature. 1982;297(5865):365–371. - PubMed
-
- Davidson EH, Jacobs HT, Britten RJ. Very short repeats and coordinate induction of genes. Nature. 1983;301(5900):468–470. - PubMed
-
- Li S, Shang Y. Regulation of SRC family coactivators by post-translational modifications. Cellular Signalling. 2007;19(6):1101–1112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
