No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1
- PMID: 22110622
- PMCID: PMC3217935
- DOI: 10.1371/journal.pone.0027266
No apparent costs for facultative antibiotic production by the soil bacterium Pseudomonas fluorescens Pf0-1
Abstract
Background: Many soil-inhabiting bacteria are known to produce secondary metabolites that can suppress microorganisms competing for the same resources. The production of antimicrobial compounds is expected to incur fitness costs for the producing bacteria. Such costs form the basis for models on the co-existence of antibiotic-producing and non-antibiotic producing strains. However, so far studies quantifying the costs of antibiotic production by bacteria are scarce. The current study reports on possible costs, for antibiotic production by Pseudomonas fluorescens Pf0-1, a soil bacterium that is induced to produce a broad-spectrum antibiotic when it is confronted with non-related bacterial competitors or supernatants of their cultures.
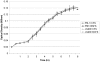
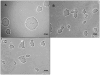
Methodology and principal findings: We measured the possible cost of antibiotic production for Pseudomonas fluorescens Pf0-1 by monitoring changes in growth rate with and without induction of antibiotic production by supernatant of a bacterial competitor, namely Pedobacter sp.. Experiments were performed in liquid as well as on semi-solid media under nutrient-limited conditions that are expected to most clearly reveal fitness costs. Our results did not reveal any significant costs for production of antibiotics by Pseudomonas fluorescens Pf0-1. Comparison of growth rates of the antibiotic-producing wild-type cells with those of non-antibiotic producing mutants did not reveal costs of antibiotic production either.
Significance: Based on our findings we propose that the facultative production of antibiotics might not be selected to mitigate metabolic costs, but instead might be advantageous because it limits the risk of competitors evolving resistance, or even the risk of competitors feeding on the compounds produced.
Conflict of interest statement
Figures

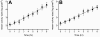
Similar articles
-
Transcriptional and antagonistic responses of Pseudomonas fluorescens Pf0-1 to phylogenetically different bacterial competitors.ISME J. 2011 Jun;5(6):973-85. doi: 10.1038/ismej.2010.196. Epub 2011 Jan 13. ISME J. 2011. PMID: 21228890 Free PMC article.
-
The effect of phylogenetically different bacteria on the fitness of Pseudomonas fluorescens in sand microcosms.PLoS One. 2015 Mar 16;10(3):e0119838. doi: 10.1371/journal.pone.0119838. eCollection 2015. PLoS One. 2015. PMID: 25774766 Free PMC article.
-
Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil.Appl Environ Microbiol. 2009 Jun;75(12):3872-81. doi: 10.1128/AEM.00017-09. Epub 2009 Apr 24. Appl Environ Microbiol. 2009. PMID: 19395572 Free PMC article.
-
The adnA transcriptional factor affects persistence and spread of Pseudomonas fluorescens under natural field conditions.Appl Environ Microbiol. 2001 Feb;67(2):852-7. doi: 10.1128/AEM.67.2.852-857.2001. Appl Environ Microbiol. 2001. PMID: 11157254 Free PMC article.
-
Exploration of Social Spreading Reveals That This Behavior Is Prevalent among Pedobacter and Pseudomonas fluorescens Isolates and That There Are Variations in the Induction of the Phenotype.Appl Environ Microbiol. 2021 Sep 10;87(19):e0134421. doi: 10.1128/AEM.01344-21. Epub 2021 Jul 21. Appl Environ Microbiol. 2021. PMID: 34288708 Free PMC article.
Cited by
-
Inhibitory interaction networks among coevolved Streptomyces populations from prairie soils.PLoS One. 2019 Oct 31;14(10):e0223779. doi: 10.1371/journal.pone.0223779. eCollection 2019. PLoS One. 2019. PMID: 31671139 Free PMC article.
-
Draft Genome Sequence of Pedobacter sp. Strain V48, Isolated from a Coastal Sand Dune in the Netherlands.Genome Announc. 2014 Feb 27;2(1):e00094-14. doi: 10.1128/genomeA.00094-14. Genome Announc. 2014. PMID: 24578271 Free PMC article.
-
Persistence in the shadow of killers.Front Microbiol. 2014 Jul 14;5:342. doi: 10.3389/fmicb.2014.00342. eCollection 2014. Front Microbiol. 2014. PMID: 25071753 Free PMC article.
-
Microbe-driven chemical ecology: past, present and future.ISME J. 2019 Nov;13(11):2656-2663. doi: 10.1038/s41396-019-0469-x. Epub 2019 Jul 9. ISME J. 2019. PMID: 31289346 Free PMC article. Review.
-
Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes.ISME J. 2014 Feb;8(2):249-56. doi: 10.1038/ismej.2013.175. Epub 2013 Oct 24. ISME J. 2014. PMID: 24152720 Free PMC article.
References
-
- Raaijmakers JM, Vlami M, de Souza JT. Antibiotic production by bacterial biocontrol agents. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 2002;81:537–547. - PubMed
-
- Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357–364. - PubMed
-
- Riley MA, Goldstone CM, Wertz JE, Gordon D. A phylogenetic approach to assessing the targets of microbial warfare. Journal of Evolutionary Biology. 2003;16:690–697. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

