Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing
- PMID: 22110666
- PMCID: PMC3217988
- DOI: 10.1371/journal.pone.0027530
Identification and characterization of microRNAs from peanut (Arachis hypogaea L.) by high-throughput sequencing
Abstract
Background: MicroRNAs (miRNAs) are noncoding RNAs of approximately 21 nt that regulate gene expression in plants post-transcriptionally by endonucleolytic cleavage or translational inhibition. miRNAs play essential roles in numerous developmental and physiological processes and many of them are conserved across species. Extensive studies of miRNAs have been done in a few model plants; however, less is known about the diversity of these regulatory RNAs in peanut (Arachis hypogaea L.), one of the most important oilseed crops cultivated worldwide.
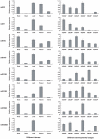
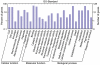
Results: A library of small RNA from peanut was constructed for deep sequencing. In addition to 126 known miRNAs from 33 families, 25 novel peanut miRNAs were identified. The miRNA* sequences of four novel miRNAs were discovered, providing additional evidence for the existence of miRNAs. Twenty of the novel miRNAs were considered to be species-specific because no homolog has been found for other plant species. qRT-PCR was used to analyze the expression of seven miRNAs in different tissues and in seed at different developmental stages and some showed tissue- and/or growth stage-specific expression. Furthermore, potential targets of these putative miRNAs were predicted on the basis of the sequence homology search.
Conclusions: We have identified large numbers of miRNAs and their related target genes through deep sequencing of a small RNA library. This study of the identification and characterization of miRNAs in peanut can initiate further study on peanut miRNA regulation mechanisms, and help toward a greater understanding of the important roles of miRNAs in peanut.
Conflict of interest statement
Figures
References
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. - PubMed
-
- Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–259. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials