EzrA contributes to the regulation of cell size in Staphylococcus aureus
- PMID: 22110668
- PMCID: PMC3215724
- DOI: 10.1371/journal.pone.0027542
EzrA contributes to the regulation of cell size in Staphylococcus aureus
Abstract
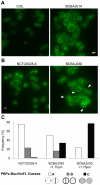
EzrA is a negative regulator of FtsZ in Bacillus subtilis, involved in the coordination between cell growth and cell division and in the control of the cell elongation-division cycle. We have now studied the role of the Staphylococcus aureus homologue of the B. subtilis EzrA protein and shown that it is not essential for cell viability. EzrA conditional and null mutants have an overall increase of the average cell size, compared to wild type strains. In the larger ezrA mutant S. aureus cells, cell division protein FtsZ and the cell wall synthesizing Penicillin Binding Proteins (PBPs) are not properly localized. This suggests that there may be a maximum cell diameter that allows formation of a Z-ring capable of recruiting the other components of the divisome and of driving cytokinesis. We propose that the major role of EzrA in S. aureus is in cell size homeostasis.
Conflict of interest statement
Figures
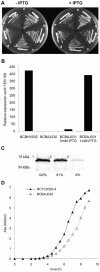
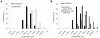

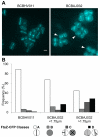
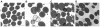


References
-
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. - PubMed
-
- Bi EF, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
-
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:R514–526. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

