Dynamic modulation of thymic microRNAs in response to stress
- PMID: 22110677
- PMCID: PMC3217971
- DOI: 10.1371/journal.pone.0027580
Dynamic modulation of thymic microRNAs in response to stress
Abstract
Physiological stress evokes rapid changes in both the innate and adaptive immune response. Immature αβ T cells developing in the thymus are particularly sensitive to stress, with infections and/or exposure to lipopolysaccharide or glucocorticoids eliciting a rapid apoptotic program. MicroRNAs are a class of small, non-coding RNAs that regulate global gene expression by targeting diverse mRNAs for degradation. We hypothesized that a subset of thymically encoded microRNAs would be stress responsive and modulate thymopoiesis. We performed microRNA profiling of thymic microRNAs isolated from control or stressed thymic tissue obtained from mice. We identified 18 microRNAs that are dysregulated >1.5-fold in response to lipopolysaccharide or the synthetic corticosteroid dexamethasone. These included the miR-17-90 cluster, which have anti-apoptotic functions, and the miR-181 family, which contribute to T cell tolerance. The stress-induced changes in the thymic microRNAs are dynamically and distinctly regulated in the CD4(-)CD8(-), CD4(+)CD8(+), CD4(+)CD8(-), and CD4(-)CD8(+) thymocyte subsets. Several of the differentially regulated murine thymic miRs are also stress responsive in the heart, kidney, liver, brain, and/or spleen. The most dramatic thymic microRNA down modulated is miR-181d, exhibiting a 15-fold reduction following stress. This miR has both similar and distinct gene targets as miR-181a, another member of miR-181 family. Many of the differentially regulated microRNAs have known functions in thymopoiesis, indicating that their dysregulation will alter T cell repertoire selection and the formation of naïve T cells. This data has implications for clinical treatments involving anti-inflammatory steroids, ablation therapies, and provides mechanistic insights into the consequences of infections.
Conflict of interest statement
Figures
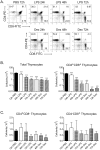
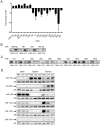
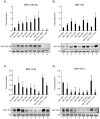
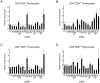

References
-
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. - PubMed
-
- Douek DC, McFarland RD, Keiser PH, Gage EA, Massey JM, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–695. - PubMed
-
- Murgita RA, Wigzell H. Regulation of immune functions in the fetus and newborn. Prog Allergy. 1981;29:54–133. - PubMed
-
- Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl):S57–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

