Evaluating genetic markers and neurobiochemical analytes for fluoxetine response using a panel of mouse inbred strains
- PMID: 22113448
- PMCID: PMC3337404
- DOI: 10.1007/s00213-011-2574-z
Evaluating genetic markers and neurobiochemical analytes for fluoxetine response using a panel of mouse inbred strains
Abstract
Rationale: Identification of biomarkers that establish diagnosis or treatment response is critical to the advancement of research and management of patients with depression.
Objective: Our goal was to identify biomarkers that can potentially assess fluoxetine response and risk to poor treatment outcome.
Methods: We measured behavior, gene expression, and the levels of 36 neurobiochemical analytes across a panel of genetically diverse mouse inbred lines after chronic treatment with water or fluoxetine.
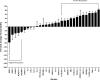
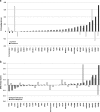
Results: Glyoxylase 1 (GLO1) and guanine nucleotide-binding protein 1 (GNB1) mostly account for baseline anxiety-like and depressive-like behavior, indicating a common biological link between depression and anxiety. Fluoxetine-induced biochemical alterations discriminated positive responders, while baseline neurobiochemical differences differentiated negative responders (p < 0.006). Results show that glial fibrillary acidic protein, S100 beta protein, GLO1, and histone deacetylase 5 contributed most to fluoxetine response. These proteins are linked within a cellular growth/proliferation pathway, suggesting the involvement of cellular genesis in fluoxetine response. Furthermore, a candidate genetic locus that associates with baseline depressive-like behavior contains a gene that encodes for cellular proliferation/adhesion molecule (Cadm1), supporting a genetic basis for the role of neuro/gliogenesis in depression.
Conclusion: We provided a comprehensive analysis of behavioral, neurobiochemical, and transcriptome data across 30 mouse inbred strains that has not been accomplished before. We identified biomarkers that influence fluoxetine response, which, altogether, implicate the importance of cellular genesis in fluoxetine treatment. More broadly, this approach can be used to assess a wide range of drug response phenotypes that are challenging to address in human samples.
Figures


Similar articles
-
Genetic regulation of behavioral and neuronal responses to fluoxetine.Neuropsychopharmacology. 2008 May;33(6):1312-22. doi: 10.1038/sj.npp.1301497. Epub 2007 Jul 4. Neuropsychopharmacology. 2008. PMID: 17609676
-
Sensitivity to the effects of pharmacologically selective antidepressants in different strains of mice.Psychopharmacology (Berl). 2001 May;155(3):315-22. doi: 10.1007/s002130100694. Psychopharmacology (Berl). 2001. PMID: 11432695
-
Behavioral destabilization induced by the selective serotonin reuptake inhibitor fluoxetine.Mol Brain. 2011 Mar 16;4:12. doi: 10.1186/1756-6606-4-12. Mol Brain. 2011. PMID: 21410937 Free PMC article.
-
MPTP-induced hippocampal effects on serotonin, dopamine, neurotrophins, adult neurogenesis and depression-like behavior are partially influenced by fluoxetine in adult mice.Brain Res. 2012 May 31;1457:51-69. doi: 10.1016/j.brainres.2012.03.046. Epub 2012 Mar 27. Brain Res. 2012. PMID: 22520437
-
The age-dependent effects of selective serotonin reuptake inhibitors in humans and rodents: A review.Prog Neuropsychopharmacol Biol Psychiatry. 2011 Aug 1;35(6):1400-8. doi: 10.1016/j.pnpbp.2010.09.013. Epub 2010 Sep 25. Prog Neuropsychopharmacol Biol Psychiatry. 2011. PMID: 20883714 Review.
Cited by
-
Determinants of host susceptibility to murine respiratory syncytial virus (RSV) disease identify a role for the innate immunity scavenger receptor MARCO gene in human infants.EBioMedicine. 2016 Sep;11:73-84. doi: 10.1016/j.ebiom.2016.08.011. Epub 2016 Aug 6. EBioMedicine. 2016. PMID: 27554839 Free PMC article.
-
Gene expression signatures of response to fluoxetine treatment: systematic review and meta-analyses.Mol Psychiatry. 2025 Jul 17. doi: 10.1038/s41380-025-03118-6. Online ahead of print. Mol Psychiatry. 2025. PMID: 40676137
-
Prenatal antidepressant exposure associated with CYP2E1 DNA methylation change in neonates.Epigenetics. 2015;10(5):361-72. doi: 10.1080/15592294.2015.1026031. Epub 2015 Apr 18. Epigenetics. 2015. PMID: 25891251 Free PMC article.
-
Effects of glucocorticoids in depression: Role of astrocytes.AIMS Neurosci. 2018 Aug 28;5(3):200-210. doi: 10.3934/Neuroscience.2018.3.200. eCollection 2018. AIMS Neurosci. 2018. PMID: 32341961 Free PMC article. Review.
-
Glyoxalase 1 increases anxiety by reducing GABAA receptor agonist methylglyoxal.J Clin Invest. 2012 Jun;122(6):2306-15. doi: 10.1172/JCI61319. Epub 2012 May 15. J Clin Invest. 2012. PMID: 22585572 Free PMC article.
References
-
- Amsterdam JD, Fawcett J, Quitkin FM, Reimherr FW, Rosenbaum JF, Michelson D, Hornig-Rohan M, Beasley CM. Fluoxetine and norfluoxetine plasma concentrations in major depression: a multicenter study. Am J Psychiatry. 1997;154:963–969. - PubMed
-
- Andreescu C, Lenze EJ, Dew MA, Begley AE, Mulsant BH, Dombrovski AY, Pollock BG, Stack J, Miller MD, Reynolds CF. Effect of comorbid anxiety on treatment response and relapse risk in late-life depression: Controlled study. Br J Psychiatry. 2007;190:344–349. doi: 10.1192/bjp.bp.106.027169. - DOI - PubMed
-
- Arai M, Yuzawa H, Nohara I, Ohnishi T, Obata N, Iwayama Y, Haga S, Toyota T, Ujike H, Ichikawa T, Nishida A, Tanaka Y, Furukawa A, Aikawa Y, Kuroda O, Niizato K, Izawa R, Nakamura K, Mori N, Matsuzawa D, Hashimoto K, Iyo M, Sora I, Matsushita M, Okazaki Y, Yoshikawa T, Miyata T, Itokawa M. Enhanced carbonyl stress in a subpopulation of schizophrenia. Arch Gen Psychiatry. 2010;67:589–597. doi: 10.1001/archgenpsychiatry.2010.62. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

