Deletion of the SPARC acidic domain or EGF-like module reduces SPARC-induced migration and signaling through p38 MAPK/HSP27 in glioma
- PMID: 22114076
- PMCID: PMC3271264
- DOI: 10.1093/carcin/bgr276
Deletion of the SPARC acidic domain or EGF-like module reduces SPARC-induced migration and signaling through p38 MAPK/HSP27 in glioma
Abstract
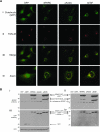
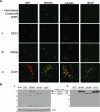
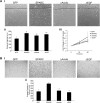
We previously demonstrated that secreted protein acidic and rich in cysteine (SPARC) increases heat shock protein 27 (HSP27) expression and phosphorylation and promotes glioma cell migration through the p38 mitogen-activated protein kinase (MAPK)/HSP27 signaling pathway. As different regions of the SPARC protein mediate different SPARC functions, elucidating which SPARC domains regulate HSP27 expression, signaling and migration might provide potential therapeutic strategies to target these functions. To investigate the roles of specific domains, we used an SPARC-green fluorescent protein (GFP) fusion protein and constructs of SPARC-GFP with deletions of either the acidic domain (ΔAcidic) or the epidermal growth factor (EGF)-like module (ΔEGF). GFP, SPARC-GFP and the two deletion mutants were expressed in U87MG glioma cells. Characterization of the derived stable clones by confocal imaging and western blotting suggests proper folding, processing and secretion of the deletion constructs. Uptake of the constructs by naive cells suggests enhanced internalization of ΔAcidic and reduced internalization of ΔEGF. Wound and transwell migration assays and western blot analysis confirm our previous results and indicate that ΔAcidic reduces SPARC-induced migration and p38 MAPK/HSP27 signaling and ΔEGF decreases SPARC-induced migration and dramatically decreases the expression and phosphorylation of HSP27 but is poorly internalized. Loss of the EGF-like module suppresses the enhanced HSP27 protein stability conferred by SPARC. In conclusion, deletions of the acidic domain and EGF-like module have differential effects on cell surface binding and HSP27 protein stability; however, both regions regulate SPARC-induced migration and signaling through HSP27. Our data link the domains of SPARC with different functions and suggest one or both of the constructs as potential therapeutic agents to inhibit SPARC-induced migration.
Figures



References
-
- Rempel SA, et al. Tumor invasiveness and anti-invasion strategies. In: Newton HB, editor. Handbook of Brain Tumor Chemotherapy. 2006. Elsevier Inc., San Diego, CA, pp. 193–218.
-
- Lefranc F, et al. Possible future issues in the treatment of glioblastomas: special emphasis on cell migration and the resistance of migrating glioblastoma cells to apoptosis. J. Clin. Oncol. 2005;23:2411–2422. - PubMed
-
- Louis DN. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. 2006;1:97–117. - PubMed
-
- Demuth T, et al. Molecular mechanisms of glioma cell migration and invasion. J. Neurooncol. 2004;70:217–228. - PubMed
-
- Sage H, et al. Characterization of a novel serum albumin-binding glycoprotein secreted by endothelial cells in culture. J. Biol. Chem. 1984;259:3993–4007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

