MetaQC: objective quality control and inclusion/exclusion criteria for genomic meta-analysis
- PMID: 22116060
- PMCID: PMC3258120
- DOI: 10.1093/nar/gkr1071
MetaQC: objective quality control and inclusion/exclusion criteria for genomic meta-analysis
Abstract
Genomic meta-analysis to combine relevant and homogeneous studies has been widely applied, but the quality control (QC) and objective inclusion/exclusion criteria have been largely overlooked. Currently, the inclusion/exclusion criteria mostly depend on ad-hoc expert opinion or naïve threshold by sample size or platform. There are pressing needs to develop a systematic QC methodology as the decision of study inclusion greatly impacts the final meta-analysis outcome. In this article, we propose six quantitative quality control measures, covering internal homogeneity of coexpression structure among studies, external consistency of coexpression pattern with pathway database, and accuracy and consistency of differentially expressed gene detection or enriched pathway identification. Each quality control index is defined as the minus log transformed P values from formal hypothesis testing. Principal component analysis biplots and a standardized mean rank are applied to assist visualization and decision. We applied the proposed method to 4 large-scale examples, combining 7 brain cancer, 9 prostate cancer, 8 idiopathic pulmonary fibrosis and 17 major depressive disorder studies, respectively. The identified problematic studies were further scrutinized for potential technical or biological causes of their lower quality to determine their exclusion from meta-analysis. The application and simulation results concluded a systematic quality assessment framework for genomic meta-analysis.
Figures



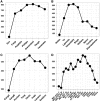
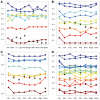
References
-
- Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

