Regulation of membrane protein degradation by starvation-response pathways
- PMID: 22118530
- PMCID: PMC3276697
- DOI: 10.1111/j.1600-0854.2011.01314.x
Regulation of membrane protein degradation by starvation-response pathways
Abstract
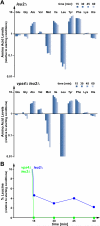
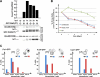
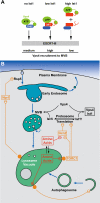
The multivesicular body (MVB) pathway delivers membrane proteins to the lumen of the vacuole/lysosome for degradation. The resulting amino acids are transported to the cytoplasm for reuse in protein synthesis. Our study shows that this amino acid recycling system plays an essential role in the adaptation of cells to starvation conditions. Cells respond to amino acid starvation by upregulating both endocytosis and the MVB pathway, thereby providing amino acids through increased protein turnover. Our data suggest that increased Rsp5-dependent ubiquitination of membrane proteins and a drop in Ist1 levels, a negative regulator of endosomal sorting complex required for transport (ESCRT) activity, cause this response. Furthermore, we found that target of rapamycin complex 1 (TORC1) and a second, unknown nutrient-sensing system are responsible for the starvation-induced protein turnover. Together, the data indicate that protein synthesis and turnover are linked by a common regulatory system that ensures adaptation and survival under nutrient-stress conditions.
© 2011 John Wiley & Sons A/S.
Figures





References
-
- Wang X, Proud CG. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009;19(6):260–267. - PubMed
-
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124(3):471–484. - PubMed
-
- Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, Neufeld TP. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans. 2009;37(Pt 1):232–236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

