Targeted enrichment of genomic DNA regions for next-generation sequencing
- PMID: 22121152
- PMCID: PMC3245553
- DOI: 10.1093/bfgp/elr033
Targeted enrichment of genomic DNA regions for next-generation sequencing
Abstract
In this review, we discuss the latest targeted enrichment methods and aspects of their utilization along with second-generation sequencing for complex genome analysis. In doing so, we provide an overview of issues involved in detecting genetic variation, for which targeted enrichment has become a powerful tool. We explain how targeted enrichment for next-generation sequencing has made great progress in terms of methodology, ease of use and applicability, but emphasize the remaining challenges such as the lack of even coverage across targeted regions. Costs are also considered versus the alternative of whole-genome sequencing which is becoming ever more affordable. We conclude that targeted enrichment is likely to be the most economical option for many years to come in a range of settings.
Figures
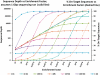
 [52]
[52]  EF, enrichment factor; ROI, region of interest in kb; genome, genome size in kb; pot, percent on target sequences; seq per run, assumed sequences per run in kb.
EF, enrichment factor; ROI, region of interest in kb; genome, genome size in kb; pot, percent on target sequences; seq per run, assumed sequences per run in kb.

Similar articles
-
Reduced representation methods for subgenomic enrichment and next-generation sequencing.Methods Mol Biol. 2011;772:85-103. doi: 10.1007/978-1-61779-228-1_5. Methods Mol Biol. 2011. PMID: 22065433 Review.
-
Targeted capture in evolutionary and ecological genomics.Mol Ecol. 2016 Jan;25(1):185-202. doi: 10.1111/mec.13304. Epub 2015 Jul 30. Mol Ecol. 2016. PMID: 26137993 Free PMC article. Review.
-
Standard enrichment methods for targeted next-generation sequencing in high-repeat genomic regions.Genet Med. 2013 Nov;15(11):910-1. doi: 10.1038/gim.2013.119. Genet Med. 2013. PMID: 24196000 No abstract available.
-
Computational methods for discovering structural variation with next-generation sequencing.Nat Methods. 2009 Nov;6(11 Suppl):S13-20. doi: 10.1038/nmeth.1374. Nat Methods. 2009. PMID: 19844226 Review.
-
Applications of next-generation sequencing to phylogeography and phylogenetics.Mol Phylogenet Evol. 2013 Feb;66(2):526-38. doi: 10.1016/j.ympev.2011.12.007. Epub 2011 Dec 14. Mol Phylogenet Evol. 2013. PMID: 22197804 Review.
Cited by
-
Regulatory function and mechanism research for m6A modification WTAP via SUCLG2-AS1- miR-17-5p-JAK1 axis in AML.BMC Cancer. 2024 Jan 17;24(1):98. doi: 10.1186/s12885-023-11687-4. BMC Cancer. 2024. PMID: 38233760 Free PMC article.
-
Application of targeted enrichment to next-generation sequencing of retroviruses integrated into the host human genome.Sci Rep. 2016 Jun 20;6:28324. doi: 10.1038/srep28324. Sci Rep. 2016. PMID: 27321866 Free PMC article.
-
A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection.Nat Commun. 2018 Nov 27;9(1):5012. doi: 10.1038/s41467-018-07324-5. Nat Commun. 2018. PMID: 30479331 Free PMC article.
-
Towards Next-Generation Sequencing (NGS)-Based Newborn Screening: A Technical Study to Prepare for the Challenges Ahead.Int J Neonatal Screen. 2022 Feb 24;8(1):17. doi: 10.3390/ijns8010017. Int J Neonatal Screen. 2022. PMID: 35323196 Free PMC article.
-
PKD1 Duplicated regions limit clinical Utility of Whole Exome Sequencing for Genetic Diagnosis of Autosomal Dominant Polycystic Kidney Disease.Sci Rep. 2019 Mar 11;9(1):4141. doi: 10.1038/s41598-019-40761-w. Sci Rep. 2019. PMID: 30858458 Free PMC article.
References
-
- Fuller CW, Middendorf LR, Benner SA, et al. The challenges of sequencing by synthesis. Nat Biotechnol. 2009;27(11):1013–23. - PubMed
-
- Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11(1):31–46. - PubMed
-
- Albert TJ, Molla MN, Muzny DM, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–5. - PubMed
-
- Bashiardes S, Veile R, Helms C, et al. Direct genomic selection. Nat Methods. 2005;2(1):63–9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

