Sequence similarity between the erythrocyte binding domain of the Plasmodium vivax Duffy binding protein and the V3 loop of HIV-1 strain MN reveals a functional heparin binding motif involved in binding to the Duffy antigen receptor for chemokines
- PMID: 22122911
- PMCID: PMC3240837
- DOI: 10.1186/1743-422X-8-523
Sequence similarity between the erythrocyte binding domain of the Plasmodium vivax Duffy binding protein and the V3 loop of HIV-1 strain MN reveals a functional heparin binding motif involved in binding to the Duffy antigen receptor for chemokines
Abstract
Background: The HIV surface glycoprotein gp120 (SU, gp120) and the Plasmodium vivax Duffy binding protein (PvDBP) bind to chemokine receptors during infection and have a site of amino acid sequence similarity in their binding domains that often includes a heparin binding motif (HBM). Infection by either pathogen has been found to be inhibited by polyanions.
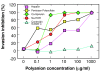
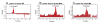
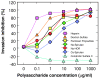
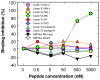
Results: Specific polyanions that inhibit HIV infection and bind to the V3 loop of X4 strains also inhibited DBP-mediated infection of erythrocytes and DBP binding to the Duffy Antigen Receptor for Chemokines (DARC). A peptide including the HBM of PvDBP had similar affinity for heparin as RANTES and V3 loop peptides, and could be specifically inhibited from heparin binding by the same polyanions that inhibit DBP binding to DARC. However, some V3 peptides can competitively inhibit RANTES binding to heparin, but not the PvDBP HBM peptide. Three other members of the DBP family have an HBM sequence that is necessary for erythrocyte binding, however only the protein which binds to DARC, the P. knowlesi alpha protein, is inhibited by heparin from binding to erythrocytes. Heparitinase digestion does not affect the binding of DBP to erythrocytes.
Conclusion: The HBMs of DBPs that bind to DARC have similar heparin binding affinities as some V3 loop peptides and chemokines, are responsible for specific sulfated polysaccharide inhibition of parasite binding and invasion of red blood cells, and are more likely to bind to negative charges on the receptor than cell surface glycosaminoglycans.
Figures



Similar articles
-
Sequence similarity between the erythrocyte binding domain 1 of the Plasmodium vivax Duffy binding protein and the V3 loop of HIV-1 strain MN reveals binding residues for the Duffy Antigen Receptor for Chemokines.Virol J. 2011 Jan 31;8:45. doi: 10.1186/1743-422X-8-45. Virol J. 2011. PMID: 21281498 Free PMC article.
-
Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC).Mol Microbiol. 2005 Mar;55(5):1413-22. doi: 10.1111/j.1365-2958.2004.04478.x. Mol Microbiol. 2005. PMID: 15720550
-
Independent Origin and Global Distribution of Distinct Plasmodium vivax Duffy Binding Protein Gene Duplications.PLoS Negl Trop Dis. 2016 Oct 31;10(10):e0005091. doi: 10.1371/journal.pntd.0005091. eCollection 2016 Oct. PLoS Negl Trop Dis. 2016. PMID: 27798646 Free PMC article.
-
Targeting the Plasmodium vivax Duffy-binding protein.Trends Parasitol. 2008 Jan;24(1):29-34. doi: 10.1016/j.pt.2007.10.004. Epub 2007 Nov 26. Trends Parasitol. 2008. PMID: 18023618 Review.
-
[Molecular basis and structure-activity relationships of the Duffy blood group antigens: chemokine and Plasmodium vivax receptors].Transfus Clin Biol. 2000 Oct;7(5):497-509. doi: 10.1016/s1246-7820(00)80038-5. Transfus Clin Biol. 2000. PMID: 11109635 Review. French.
Cited by
-
Blood Group Antigens C, Lub and P1 May Have a Role in HIV Infection in Africans.PLoS One. 2016 Feb 22;11(2):e0149883. doi: 10.1371/journal.pone.0149883. eCollection 2016. PLoS One. 2016. PMID: 26900853 Free PMC article.
-
Pathogenesis of HIV-associated nephropathy in children and adolescents: taking a hard look 40 years later in the era of gene-environment interactions.Am J Physiol Renal Physiol. 2024 Dec 1;327(6):F1049-F1066. doi: 10.1152/ajprenal.00208.2024. Epub 2024 Sep 26. Am J Physiol Renal Physiol. 2024. PMID: 39323389 Review.
-
The role of red blood cells in enhancing or preventing HIV infection and other diseases.Biomed Res Int. 2013;2013:758682. doi: 10.1155/2013/758682. Epub 2013 Oct 10. Biomed Res Int. 2013. PMID: 24224178 Free PMC article. Review.
References
-
- Briggs TDLSJWGMM. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages) Aids. 2000. - PubMed
-
- Shimizu N, Haraguchi Y, Takeuchi Y, Soda Y, Kanbe K, Hoshino H. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the env protein. Virol. 1999;259:324–333. doi: 10.1006/viro.1999.9764. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

