A pre-initiation complex at the 3'-end of genes drives antisense transcription independent of divergent sense transcription
- PMID: 22123739
- PMCID: PMC3315312
- DOI: 10.1093/nar/gkr1121
A pre-initiation complex at the 3'-end of genes drives antisense transcription independent of divergent sense transcription
Abstract
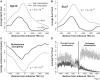
The precise nature of antisense transcripts in eukaryotes such as Saccharomyces cerevisiae remains elusive. Here we show that the 3' regions of genes possess a promoter architecture, including a pre-initiation complex (PIC), which mirrors that at the 5' region and which is much more pronounced at genes with a defined antisense transcript. Remarkably, for genes with an antisense transcript, average levels of PIC components at the 3' region are ∼60% of those at the 5' region. Moreover, at these genes, average levels of nascent antisense transcription are ∼45% of sense transcription. We find that this 3' promoter architecture persists for highly transcribed antisense transcripts where there are only low levels of transcription in the divergent sense direction, suggesting that the 3' regions of genes can drive antisense transcription independent of divergent sense transcription. To validate this, we insert short 3' regions into the middle of other genes and find that they are capable of both initiating antisense transcripts and terminating sense transcripts. Our results suggest that antisense transcription can be regulated independently of divergent sense transcription in a PIC-dependent manner and we propose that regulated production of antisense transcripts represents a fundamental and widespread component of gene regulation.
Figures






References
-
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. - PubMed
-
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. - PubMed
-
- Camblong J, Iglesias N, Fickentscher F, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

