Genome-wide antisense transcription drives mRNA processing in bacteria
- PMID: 22123973
- PMCID: PMC3250193
- DOI: 10.1073/pnas.1113521108
Genome-wide antisense transcription drives mRNA processing in bacteria
Abstract
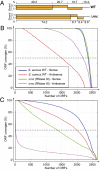
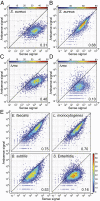
RNA deep sequencing technologies are revealing unexpected levels of complexity in bacterial transcriptomes with the discovery of abundant noncoding RNAs, antisense RNAs, long 5' and 3' untranslated regions, and alternative operon structures. Here, by applying deep RNA sequencing to both the long and short RNA fractions (<50 nucleotides) obtained from the major human pathogen Staphylococcus aureus, we have detected a collection of short RNAs that is generated genome-wide through the digestion of overlapping sense/antisense transcripts by RNase III endoribonuclease. At least 75% of sense RNAs from annotated genes are subject to this mechanism of antisense processing. Removal of RNase III activity reduces the amount of short RNAs and is accompanied by the accumulation of discrete antisense transcripts. These results suggest the production of pervasive but hidden antisense transcription used to process sense transcripts by means of creating double-stranded substrates. This process of RNase III-mediated digestion of overlapping transcripts can be observed in several evolutionarily diverse Gram-positive bacteria and is capable of providing a unique genome-wide posttranscriptional mechanism to adjust mRNA levels.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Selinger DW, et al. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. - PubMed
-
- Güell M, et al. Transcriptome complexity in a genome-reduced bacterium. Science. 2009;326:1268–1271. - PubMed
-
- Toledo-Arana A, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–956. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

