Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells
- PMID: 22124016
- PMCID: PMC3261741
- DOI: 10.1261/rna.030106.111
Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells
Abstract
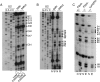
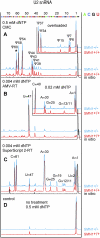
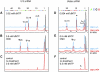
The survival of motor neuron (SMN) protein plays an important role in the biogenesis of spliceosomal snRNPs and is one factor required for the integrity of nuclear Cajal bodies (CBs). CBs are enriched in small CB-specific (sca) RNAs, which guide the formation of pseudouridylated and 2'-O-methylated residues in the snRNAs. Because SMN-deficient cells lack typical CBs, we asked whether the modification of internal residues of major and minor snRNAs is defective in these cells. We mapped modified nucleotides in the major U2 and the minor U4atac and U12 snRNAs. Using both radioactive and fluorescent primer extension approaches, we found that modification of major and minor spliceosomal snRNAs is normal in SMN-deficient cells. Our experiments also revealed a previously undetected pseudouridine at position 60 in human U2 and 2'-O-methylation of A1, A2, and G19 in human U4atac. These results confirm, and extend to minor snRNAs, previous experiments showing that scaRNPs can function in the absence of typical CBs. Furthermore, they show that the differential splicing defects in SMN-deficient cells are not due to failure of post-transcriptional modification of either major or minor snRNAs.
Figures
References
-
- Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonné R 2011. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum Mol Genet 20: 641–648 - PubMed
-
- Cioce M, Lamond A 2005. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131 - PubMed
-
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH 1997. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 6: 1205–1214 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
