Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation
- PMID: 22124152
- PMCID: PMC3266600
- DOI: 10.1128/MCB.06570-11
Genetic reevaluation of the role of F-box proteins in cyclin D1 degradation
Abstract
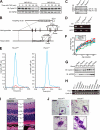
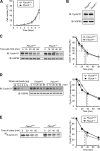
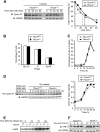
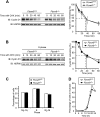
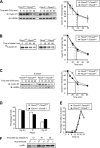
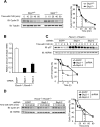
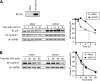
D-type cyclins play a pivotal role in G(1)-S progression of the cell cycle, and their expression is frequently deregulated in cancer. Cyclin D1 has a half-life of only ~30 min as a result of its ubiquitylation and proteasomal degradation, with various F-box proteins, including Fbxo4, Fbxw8, Skp2, and Fbxo31, having been found to contribute to its ubiquitylation. We have now generated Fbxo4-deficient mice and found no abnormalities in these animals. Cyclin D1 accumulation was thus not observed in Fbxo4(-/-) mouse tissues. The half-life of cyclin D1 in mouse embryonic fibroblasts (MEFs) prepared from Fbxo4(-/-), Fbxw8(-/-), and Fbxo4(-/-); Fbxw8(-/-) mice also did not differ from that in wild-type MEFs. Additional depletion of Skp2 and Fbxo31 in Fbxo4(-/-); Fbxw8(-/-) MEFs by RNA interference did not affect cyclin D1 stability. Although Fbxo31 depletion in MEFs increased cyclin D1 abundance, this effect appeared attributable to upregulation of cyclin D1 mRNA. Furthermore, abrogation of the function of the Skp1-Cul1-F-box protein (SCF) complex or the anaphase-promoting complex/cyclosome (APC/C) complexes did not alter the half-life of cyclin D1, whereas cyclin D1 degradation was dependent largely on proteasome activity. Our genetic analyses thus do not support a role for any of the four F-box proteins examined in cyclin D1 degradation during normal cell cycle progression. They suggest the existence of other ubiquitin ligases that target cyclin D1 for proteolysis.
Figures




References
-
- Agami R, Bernards R. 2000. Distinct initiation and maintenance mechanisms cooperate to induce G1 cell cycle arrest in response to DNA damage. Cell 102:55–66 - PubMed
-
- Ang XL, Wade Harper J. 2005. SCF-mediated protein degradation and cell cycle control. Oncogene 24:2860–2870 - PubMed
-
- Baldin V, Lukas J, Marcote MJ, Pagano M, Draetta G. 1993. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7:812–821 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
