ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites
- PMID: 22124328
- PMCID: PMC3273392
- DOI: 10.1038/emboj.2011.429
ACBD3-mediated recruitment of PI4KB to picornavirus RNA replication sites
Abstract
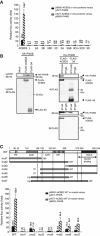
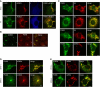
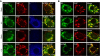
Phosphatidylinositol 4-kinase IIIβ (PI4KB) is a host factor required for genome RNA replication of enteroviruses, small non-enveloped viruses belonging to the family Picornaviridae. Here, we demonstrated that PI4KB is also essential for genome replication of another picornavirus, Aichi virus (AiV), but is recruited to the genome replication sites by a different strategy from that utilized by enteroviruses. AiV non-structural proteins, 2B, 2BC, 2C, 3A, and 3AB, interacted with a Golgi protein, acyl-coenzyme A binding domain containing 3 (ACBD3). Furthermore, we identified previously unknown interaction between ACBD3 and PI4KB, which provides a novel manner of Golgi recruitment of PI4KB. Knockdown of ACBD3 or PI4KB suppressed AiV RNA replication. The viral proteins, ACBD3, PI4KB, and phophatidylinositol-4-phosphate (PI4P) localized to the viral RNA replication sites. AiV replication and recruitment of PI4KB to the RNA replication sites were not affected by brefeldin A, in contrast to those in enterovirus infection. These results indicate that a viral protein/ACBD3/PI4KB complex is formed to synthesize PI4P at the AiV RNA replication sites and plays an essential role in viral RNA replication.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
A complex comprising phosphatidylinositol 4-kinase IIIβ, ACBD3, and Aichi virus proteins enhances phosphatidylinositol 4-phosphate synthesis and is critical for formation of the viral replication complex.J Virol. 2014 Jun;88(12):6586-98. doi: 10.1128/JVI.00208-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672044 Free PMC article.
-
Model of OSBP-Mediated Cholesterol Supply to Aichi Virus RNA Replication Sites Involving Protein-Protein Interactions among Viral Proteins, ACBD3, OSBP, VAP-A/B, and SAC1.J Virol. 2018 Mar 28;92(8):e01952-17. doi: 10.1128/JVI.01952-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29367253 Free PMC article.
-
ACBD3 Is an Essential Pan-enterovirus Host Factor That Mediates the Interaction between Viral 3A Protein and Cellular Protein PI4KB.mBio. 2019 Feb 12;10(1):e02742-18. doi: 10.1128/mBio.02742-18. mBio. 2019. PMID: 30755512 Free PMC article.
-
Emerging Role for Acyl-CoA Binding Domain Containing 3 at Membrane Contact Sites During Viral Infection.Front Microbiol. 2020 Apr 8;11:608. doi: 10.3389/fmicb.2020.00608. eCollection 2020. Front Microbiol. 2020. PMID: 32322249 Free PMC article. Review.
-
Picornavirus--host interactions to construct viral secretory membranes.Prog Mol Biol Transl Sci. 2015;129:189-212. doi: 10.1016/bs.pmbts.2014.10.007. Epub 2014 Dec 8. Prog Mol Biol Transl Sci. 2015. PMID: 25595805 Review.
Cited by
-
Purine analogs as phosphatidylinositol 4-kinase IIIβ inhibitors.Bioorg Med Chem Lett. 2016 Jun 1;26(11):2706-12. doi: 10.1016/j.bmcl.2016.04.002. Epub 2016 Apr 5. Bioorg Med Chem Lett. 2016. PMID: 27090557 Free PMC article.
-
The Amino Acid Substitution Q65H in the 2C Protein of Swine Vesicular Disease Virus Confers Resistance to Golgi Disrupting Drugs.Front Microbiol. 2016 Apr 27;7:612. doi: 10.3389/fmicb.2016.00612. eCollection 2016. Front Microbiol. 2016. PMID: 27199941 Free PMC article.
-
Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein.PLoS Pathog. 2018 May 21;14(5):e1007086. doi: 10.1371/journal.ppat.1007086. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29782554 Free PMC article.
-
New small-molecule inhibitors effectively blocking picornavirus replication.J Virol. 2014 Oct;88(19):11091-107. doi: 10.1128/JVI.01877-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008939 Free PMC article.
-
Association of NMT2 with the acyl-CoA carrier ACBD6 protects the N-myristoyltransferase reaction from palmitoyl-CoA.J Lipid Res. 2016 Feb;57(2):288-98. doi: 10.1194/jlr.M065003. Epub 2015 Nov 30. J Lipid Res. 2016. PMID: 26621918 Free PMC article.
References
-
- Aldabe R, Barco A, Carrasco L (1996) Membrane permeabilization by poliovirus proteins 2B and 2BC. J Biol Chem 271: 23134–23137 - PubMed
-
- Arita M, Nagata N, Sata T, Miyamura T, Shimizu H (2006) Quantitative analysis of poliomyelitis-like paralysis in mice induced by a poliovirus replicon. J Gen Virol 87: 3317–3327 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

