Emerging biomedical applications of synthetic biology
- PMID: 22124480
- PMCID: PMC7097403
- DOI: 10.1038/nrg3094
Emerging biomedical applications of synthetic biology
Abstract
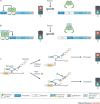
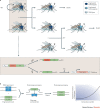
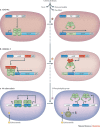
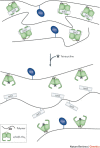
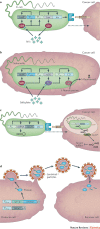
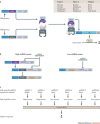
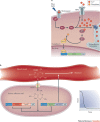
Synthetic biology aims to create functional devices, systems and organisms with novel and useful functions on the basis of catalogued and standardized biological building blocks. Although they were initially constructed to elucidate the dynamics of simple processes, designed devices now contribute to the understanding of disease mechanisms, provide novel diagnostic tools, enable economic production of therapeutics and allow the design of novel strategies for the treatment of cancer, immune diseases and metabolic disorders, such as diabetes and gout, as well as a range of infectious diseases. In this Review, we cover the impact and potential of synthetic biology for biomedical applications.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

