Investigation of VUV Photodissociation Propensities Using Peptide Libraries
- PMID: 22125417
- PMCID: PMC3224043
- DOI: 10.1016/j.ijms.2011.04.008
Investigation of VUV Photodissociation Propensities Using Peptide Libraries
Abstract
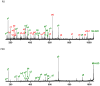
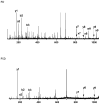
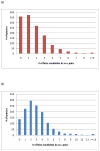
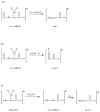
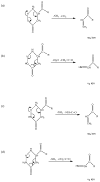
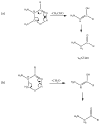
PSD does not usually generate a complete series of y-type ions, particularly at high mass, and this is a limitation for de novo sequencing algorithms. It is demonstrated that b(2) and b(3) ions can be used to help assign high mass x(N-2) and x(N-3) fragments that are found in vacuum ultraviolet (VUV) photofragmentation experiments. In addition, v(N)-type ion fragments with side chain loss from the N-terminal residue often enable confirmation of N-terminal amino acids. Libraries containing several thousand peptides were examined using photodissociation in a MALDI-TOF/TOF instrument. 1345 photodissociation spectra with a high S/N ratio were interpreted.
Figures

Similar articles
-
Peptide photodissociation at 157 nm in a linear ion trap mass spectrometer.Rapid Commun Mass Spectrom. 2005;19(12):1657-65. doi: 10.1002/rcm.1969. Rapid Commun Mass Spectrom. 2005. PMID: 15915476
-
Occurrence of C-terminal residue exclusion in peptide fragmentation by ESI and MALDI tandem mass spectrometry.J Am Soc Mass Spectrom. 2012 Feb;23(2):330-46. doi: 10.1007/s13361-011-0254-1. Epub 2011 Nov 18. J Am Soc Mass Spectrom. 2012. PMID: 22095165
-
UVnovo: A de Novo Sequencing Algorithm Using Single Series of Fragment Ions via Chromophore Tagging and 351 nm Ultraviolet Photodissociation Mass Spectrometry.Anal Chem. 2016 Apr 5;88(7):3990-7. doi: 10.1021/acs.analchem.6b00261. Epub 2016 Mar 14. Anal Chem. 2016. PMID: 26938041 Free PMC article.
-
Peptide sequencing by MALDI 193-nm photodissociation TOF MS.Methods Enzymol. 2005;402:186-209. doi: 10.1016/S0076-6879(05)02006-9. Methods Enzymol. 2005. PMID: 16401510 Review.
-
State-to-state spectroscopy and dynamics of ions and neutrals by photoionization and photoelectron methods.Annu Rev Phys Chem. 2014;65:197-224. doi: 10.1146/annurev-physchem-040412-110002. Epub 2013 Dec 13. Annu Rev Phys Chem. 2014. PMID: 24328445 Review.
Cited by
-
157 nm photodissociation of dipeptide ions containing N-terminal arginine.J Am Soc Mass Spectrom. 2014 Feb;25(2):196-203. doi: 10.1007/s13361-013-0762-2. Epub 2013 Dec 6. J Am Soc Mass Spectrom. 2014. PMID: 24310819
-
Photodissociation mass spectrometry: new tools for characterization of biological molecules.Chem Soc Rev. 2014 Apr 21;43(8):2757-83. doi: 10.1039/c3cs60444f. Epub 2014 Jan 30. Chem Soc Rev. 2014. PMID: 24481009 Free PMC article. Review.
-
Ion Activation Methods for Peptides and Proteins.Anal Chem. 2016 Jan 5;88(1):30-51. doi: 10.1021/acs.analchem.5b04563. Epub 2015 Dec 11. Anal Chem. 2016. PMID: 26630359 Free PMC article. Review.
-
Factors Affecting the Production of Aromatic Immonium Ions in MALDI 157 nm Photodissociation Studies.J Am Soc Mass Spectrom. 2016 May;27(5):834-46. doi: 10.1007/s13361-015-1329-1. Epub 2016 Feb 29. J Am Soc Mass Spectrom. 2016. PMID: 26926443
-
Enhancing Biomolecule Analysis and 2DMS Experiments by Implementation of (Activated Ion) 193 nm UVPD on a FT-ICR Mass Spectrometer.Anal Chem. 2022 Nov 15;94(45):15631-15638. doi: 10.1021/acs.analchem.2c02354. Epub 2022 Nov 1. Anal Chem. 2022. PMID: 36317856 Free PMC article.
References
-
- Aebersold R, Goodlett DR. Mass spectrometry in proteomics. Chem Rev. 2001;101(2):269–295. - PubMed
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. - PubMed
-
- Yates JR. Mass spectrometry and the age of the proteome. J Mass Spectrom. 1998;33(1):1–19. - PubMed
-
- Biemann K, Scoble HA. Characterization by tandem mass spectrometry of structural modifications in proteins. Science. 1987;237(4818):992–998. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
