PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer
- PMID: 22125492
- PMCID: PMC3219601
- DOI: 10.1371/journal.pgen.1002368
PBX1 genomic pioneer function drives ERα signaling underlying progression in breast cancer
Abstract
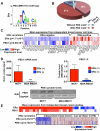
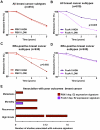
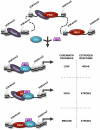
Altered transcriptional programs are a hallmark of diseases, yet how these are established is still ill-defined. PBX1 is a TALE homeodomain protein involved in the development of different types of cancers. The estrogen receptor alpha (ERα) is central to the development of two-thirds of all breast cancers. Here we demonstrate that PBX1 acts as a pioneer factor and is essential for the ERα-mediated transcriptional response driving aggressive tumors in breast cancer. Indeed, PBX1 expression correlates with ERα in primary breast tumors, and breast cancer cells depleted of PBX1 no longer proliferate following estrogen stimulation. Profiling PBX1 recruitment and chromatin accessibility across the genome of breast cancer cells through ChIP-seq and FAIRE-seq reveals that PBX1 is loaded and promotes chromatin openness at specific genomic locations through its capacity to read specific epigenetic signatures. Accordingly, PBX1 guides ERα recruitment to a specific subset of sites. Expression profiling studies demonstrate that PBX1 controls over 70% of the estrogen response. More importantly, the PBX1-dependent transcriptional program is associated with poor-outcome in breast cancer patients. Correspondingly, PBX1 expression alone can discriminate a priori the outcome in ERα-positive breast cancer patients. These features are markedly different from the previously characterized ERα-associated pioneer factor FoxA1. Indeed, PBX1 is the only pioneer factor identified to date that discriminates outcome such as metastasis in ERα-positive breast cancer patients. Together our results reveal that PBX1 is a novel pioneer factor defining aggressive ERα-positive breast tumors, as it guides ERα genomic activity to unique genomic regions promoting a transcriptional program favorable to breast cancer progression.
Conflict of interest statement
Patent pending for the use of PBX1 as a biomarker in breast cancer.
Figures




Comment in
-
Breast cancer: Staking a claim.Nat Rev Cancer. 2011 Dec 15;12(1):4. doi: 10.1038/nrc3192. Nat Rev Cancer. 2011. PMID: 22169975 No abstract available.
References
-
- Barrero MJ, Boue S, Izpisua Belmonte JC. Epigenetic mechanisms that regulate cell identity. Cell Stem Cell. 2010;7:565–570. - PubMed
-
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nature Biotechnology. 2010;28:1079–1088. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

