Gut microbiota of healthy and malnourished children in bangladesh
- PMID: 22125551
- PMCID: PMC3221396
- DOI: 10.3389/fmicb.2011.00228
Gut microbiota of healthy and malnourished children in bangladesh
Abstract
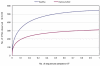
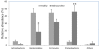
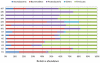
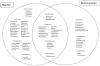
Poor health and malnutrition in preschool children are longstanding problems in Bangladesh. Gut microbiota plays a tremendous role in nutrient absorption and determining the state of health. In this study, metagenomic tool was employed to assess the gut microbiota composition of healthy and malnourished children. DNA was extracted from fecal samples of seven healthy and seven malnourished children (n = 14; age 2-3 years) were analyzed for the variable region of 16S rRNA genes by universal primer PCR followed by high-throughput 454 parallel sequencing to identify the bacterial phyla and genera. Our results reveal that the healthy children had a significantly higher number of operational taxonomic unit in their gut than that of the malnourished children (healthy vs. malnourished: 546 vs. 310). In malnourished children, bacterial population of the phyla Proteobacteria and Bacteroidetes accounted for 46 and 18%, respectively. Conversely, in healthy children, Proteobacteria and Bacteroidetes accounted for 5% and 44, respectively (p < 0.001). In malnourished children, the phylum Proteobacteria included pathogenic genera, namely Klebsiella and Escherichia, which were 174-fold and 9-fold higher, respectively, than their healthy counterpart. The predominance of potentially pathogenic Proteobacteria and minimal level of Bacteroidetes as commensal microbiota might be associated to the ill health of malnourished children in Bangladesh.
Keywords: 16S rDNA; children; gut; microbiota; nutrition.
Figures
References
-
- Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D. R., Fernandes G. R., Tap J., Bruls T., Batto J. M., Bertalan M., Borruel N., Casellas F., Fernandez L., Gautier L., Hansen T., Hattori M., Hayashi T., Kleerebezem M., Kurokawa K., Leclerc M., Levenez F., Manichanh C., Nielsen H. B., Nielsen T., Pons N., Poulain J., Qin J., Sicheritz-Ponten T., Tims S., Torrents D., Ugarte E., Zoetendal E. G., Wang J., Guarner F., Pedersen O., de Vos W. M., Brunak S., Doré J., MetaHIT Consortium. Antolín M., Artiguenave F., Blottiere H. M., Almeida M., Brechot C., Cara C., Chervaux C., Cultrone A., Delorme C., Denariaz G., Dervyn R., Foerstner K. U., Friss C., van de Guchte M., Guedon E., Haimet F., Huber W., van Hylckama-Vlieg J., Jamet A., Juste C., Kaci G., Knol J., Lakhdari O., Layec S., Le Roux K., Maguin E., Mérieux A., Melo Minardi R., M’rini C., Muller J., Oozeer R., Parkhill J., Renault P., Rescigno M., Sanchez N., Sunagawa S., Torrejon A., Turner K., Vandemeulebrouck G., Varela E., Winogradsky Y., Zeller G., Weissenbach J., Ehrlich S. D., Bork P. (2011). Enterotypes of the human gut microbiome. Nature 473, 174 –180.10.1038/nature09944 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Medical

