The proteomics of colorectal cancer: identification of a protein signature associated with prognosis
- PMID: 22125622
- PMCID: PMC3220687
- DOI: 10.1371/journal.pone.0027718
The proteomics of colorectal cancer: identification of a protein signature associated with prognosis
Abstract
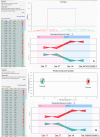
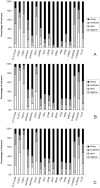
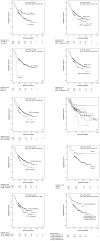
Colorectal cancer is one of the commonest types of cancer and there is requirement for the identification of prognostic biomarkers. In this study protein expression profiles have been established for colorectal cancer and normal colonic mucosa by proteomics using a combination of two dimensional gel electrophoresis with fresh frozen sections of paired Dukes B colorectal cancer and normal colorectal mucosa (n = 28), gel image analysis and high performance liquid chromatography-tandem mass spectrometry. Hierarchical cluster analysis and principal components analysis showed that the protein expression profiles of colorectal cancer and normal colonic mucosa clustered into distinct patterns of protein expression. Forty-five proteins were identified as showing at least 1.5 times increased expression in colorectal cancer and the identity of these proteins was confirmed by liquid chromatography-tandem mass spectrometry. Fifteen proteins that showed increased expression were validated by immunohistochemistry using a well characterised colorectal cancer tissue microarray containing 515 primary colorectal cancer, 224 lymph node metastasis and 50 normal colonic mucosal samples. The proteins that showed the greatest degree of overexpression in primary colorectal cancer compared with normal colonic mucosa were heat shock protein 60 (p<0.001), S100A9 (p<0.001) and translationally controlled tumour protein (p<0.001). Analysis of proteins individually identified 14-3-3β as a prognostic biomarker (χ² = 6.218, p = 0.013, HR = 0.639, 95%CI 0.448-0.913). Hierarchical cluster analysis identified distinct phenotypes associated with survival and a two-protein signature consisting of 14-3-3β and aldehyde dehydrogenase 1 was identified as showing prognostic significance (χ² = 7.306, p = 0.007, HR = 0.504, 95%CI 0.303-0.838) and that remained independently prognostic (p = 0.01, HR = 0.416, 95%CI 0.208-0.829) in a multivariate model.
Conflict of interest statement
Figures




Similar articles
-
Proteomic profiling of proteins associated with lymph node metastasis in colorectal cancer.J Cell Biochem. 2010 Aug 15;110(6):1512-9. doi: 10.1002/jcb.22726. J Cell Biochem. 2010. PMID: 20524204
-
Proteome analysis and tissue microarray for profiling protein markers associated with lymph node metastasis in colorectal cancer.J Proteome Res. 2007 Jul;6(7):2495-501. doi: 10.1021/pr060644r. Epub 2007 Jun 2. J Proteome Res. 2007. PMID: 17542627
-
Identification and Verification of Two Novel Differentially Expressed Proteins from Non-neoplastic Mucosa and Colorectal Carcinoma Via iTRAQ Combined with Liquid Chromatography-Mass Spectrometry.Pathol Oncol Res. 2020 Apr;26(2):967-976. doi: 10.1007/s12253-019-00651-y. Epub 2019 Mar 29. Pathol Oncol Res. 2020. PMID: 30927204
-
The cancer secretome, current status and opportunities in the lung, breast and colorectal cancer context.Biochim Biophys Acta. 2013 Nov;1834(11):2242-58. doi: 10.1016/j.bbapap.2013.01.029. Epub 2013 Jan 31. Biochim Biophys Acta. 2013. PMID: 23376433 Review.
-
Proteomics for discovery of candidate colorectal cancer biomarkers.World J Gastroenterol. 2014 Apr 14;20(14):3804-24. doi: 10.3748/wjg.v20.i14.3804. World J Gastroenterol. 2014. PMID: 24744574 Free PMC article. Review.
Cited by
-
Proteomic-Based Approaches for the Study of Cytokines in Lung Cancer.Dis Markers. 2016;2016:2138627. doi: 10.1155/2016/2138627. Epub 2016 Jun 30. Dis Markers. 2016. PMID: 27445423 Free PMC article. Review.
-
Proteins are potent biomarkers to detect colon cancer progression.Saudi J Biol Sci. 2017 Sep;24(6):1212-1221. doi: 10.1016/j.sjbs.2014.09.017. Epub 2014 Oct 5. Saudi J Biol Sci. 2017. PMID: 28855814 Free PMC article.
-
Prognostic Value of Cancer Stem Cell Marker ALDH1 Expression in Colorectal Cancer: A Systematic Review and Meta-Analysis.PLoS One. 2015 Dec 18;10(12):e0145164. doi: 10.1371/journal.pone.0145164. eCollection 2015. PLoS One. 2015. PMID: 26682730 Free PMC article.
-
Challenges in Biomarker Discovery: Combining Expert Insights with Statistical Analysis of Complex Omics Data.Expert Opin Med Diagn. 2013 Jan;7(1):37-51. doi: 10.1517/17530059.2012.718329. Expert Opin Med Diagn. 2013. PMID: 23335946 Free PMC article.
-
Low Expression of 14-3-3beta Is Associated With Adverse Survival of Diffuse Large B-Cell Lymphoma Patients.Front Med (Lausanne). 2019 Oct 30;6:237. doi: 10.3389/fmed.2019.00237. eCollection 2019. Front Med (Lausanne). 2019. PMID: 31737636 Free PMC article.
References
-
- Cunningham D, Atkin W, Heinz-Josef L, Lynch HT, Minsky B, et al. Colorectal cancer. Lancet. 2010;375:1030–1047. - PubMed
-
- Søreide K, Nedrebø BS, Knapp JC, Glomsaker TB, Søreide JA, et al. Evolving molecular classification by genomic and proteomic biomarkers in colorectal cancer: potential implications for the surgical oncologist. Surg Oncol. 2009;18:31–50. - PubMed
-
- Kulasingham V, Diamandis E. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5:588–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

