The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes
- PMID: 22125637
- PMCID: PMC3220699
- DOI: 10.1371/journal.pone.0027909
The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes
Abstract
Members of the genus Xenorhabdus are entomopathogenic bacteria that associate with nematodes. The nematode-bacteria pair infects and kills insects, with both partners contributing to insect pathogenesis and the bacteria providing nutrition to the nematode from available insect-derived nutrients. The nematode provides the bacteria with protection from predators, access to nutrients, and a mechanism of dispersal. Members of the bacterial genus Photorhabdus also associate with nematodes to kill insects, and both genera of bacteria provide similar services to their different nematode hosts through unique physiological and metabolic mechanisms. We posited that these differences would be reflected in their respective genomes. To test this, we sequenced to completion the genomes of Xenorhabdus nematophila ATCC 19061 and Xenorhabdus bovienii SS-2004. As expected, both Xenorhabdus genomes encode many anti-insecticidal compounds, commensurate with their entomopathogenic lifestyle. Despite the similarities in lifestyle between Xenorhabdus and Photorhabdus bacteria, a comparative analysis of the Xenorhabdus, Photorhabdus luminescens, and P. asymbiotica genomes suggests genomic divergence. These findings indicate that evolutionary changes shaped by symbiotic interactions can follow different routes to achieve similar end points.
Conflict of interest statement
Figures


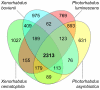
References
-
- Spratt BG, Staley JT, Fischer MC. Introduction: species and speciation in microorganisms. Phil Trans Roy Soc B. 2006;361:1897.
-
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. - PubMed
-
- Gupta RS, Griffiths E. Critical issues in bacterial phylogeny. Theor Popul Biol. 2002;61:423–434. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

