Molecular docking of 2-(benzimidazol-2-ylthio)-N-phenylacetamide-derived small-molecule agonists of human formyl peptide receptor 1
- PMID: 22127612
- PMCID: PMC3314724
- DOI: 10.1007/s00894-011-1307-x
Molecular docking of 2-(benzimidazol-2-ylthio)-N-phenylacetamide-derived small-molecule agonists of human formyl peptide receptor 1
Abstract
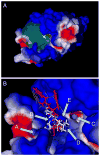
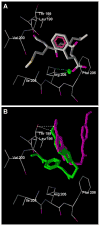
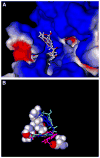
Human N-formyl peptide receptor 1 (FPR1) is a G protein-coupled receptor (GPCR) involved in host defense and sensing cellular damage. Since structure-based ligand design for many GPCRs, including FPR1, is restricted by the lack of experimental three dimensional structures, homology modeling has been widely used to study GPCR-ligand binding. Indeed, receptor-ligand binding mode predictions can be derived from homology modeling with supporting ligand information. In the present work, we report comparative docking studies of 2-(benzimidazol-2-ylthio)-N-phenylacetamide derived FPR1 agonists, identified here and previously, with several known FPR1 peptide agonists in a FPR1 homology model that is based on the crystal structure of bovine rhodopsin. We found that the binding pocket of the most active molecules shares some common features with high affinity FPR1 peptide agonists, suggesting that they may bind to similar binding sites. Classification tree analysis led to the derivation of a good recognition model based on four amino acid descriptors for distinguishing FPR1 ligands from inactive analogs. Hence, the corresponding residues (Thr199, Arg201, Gly202, and Ala261) can be considered as markers of important areas in the ligand binding site. Concurrently, we identified several unique binding features of benzimidazole derivatives and showed that alkoxy-substituents of the benzimidazole ring are located within a FPR1 hole bounded by Thr199, Thr265, Ile268, and Leu271 or in a groove in the vicinity of Leu198, Arg201, Gly202, and Arg205. The understanding of these molecular features will likely prove beneficial in future design of novel FPR1 agonists based on the benzimidazole scaffold.
Figures

Similar articles
-
The role of water in activation mechanism of human N-formyl peptide receptor 1 (FPR1) based on molecular dynamics simulations.PLoS One. 2012;7(11):e47114. doi: 10.1371/journal.pone.0047114. Epub 2012 Nov 26. PLoS One. 2012. PMID: 23189124 Free PMC article.
-
Identification of novel small-molecule agonists for human formyl peptide receptors and pharmacophore models of their recognition.Mol Pharmacol. 2010 Feb;77(2):159-70. doi: 10.1124/mol.109.060673. Epub 2009 Nov 10. Mol Pharmacol. 2010. PMID: 19903830 Free PMC article.
-
Non-peptide ligand binding to the formyl peptide receptor FPR2--A comparison to peptide ligand binding modes.Bioorg Med Chem. 2015 Jul 15;23(14):4072-81. doi: 10.1016/j.bmc.2015.03.062. Epub 2015 Mar 28. Bioorg Med Chem. 2015. PMID: 25882522
-
Basic characteristics of the neutrophil receptors that recognize formylated peptides, a danger-associated molecular pattern generated by bacteria and mitochondria.Biochem Pharmacol. 2016 Aug 15;114:22-39. doi: 10.1016/j.bcp.2016.04.014. Epub 2016 Apr 27. Biochem Pharmacol. 2016. PMID: 27131862 Review.
-
Development of small molecule non-peptide formyl peptide receptor (FPR) ligands and molecular modeling of their recognition.Curr Med Chem. 2014;21(13):1478-504. doi: 10.2174/0929867321666131218095521. Curr Med Chem. 2014. PMID: 24350845 Free PMC article. Review.
Cited by
-
3-(1H-indol-3-yl)-2-[3-(4-nitrophenyl)ureido]propanamide enantiomers with human formyl-peptide receptor agonist activity: molecular modeling of chiral recognition by FPR2.Biochem Pharmacol. 2013 Feb 1;85(3):404-16. doi: 10.1016/j.bcp.2012.11.015. Epub 2012 Dec 3. Biochem Pharmacol. 2013. PMID: 23219934 Free PMC article.
-
Antagonism of human formyl peptide receptor 1 with natural compounds and their synthetic derivatives.Int Immunopharmacol. 2016 Aug;37:43-58. doi: 10.1016/j.intimp.2015.08.036. Epub 2015 Sep 15. Int Immunopharmacol. 2016. PMID: 26382576 Free PMC article. Review.
-
Further studies on 2-arylacetamide pyridazin-3(2H)-ones: design, synthesis and evaluation of 4,6-disubstituted analogs as formyl peptide receptors (FPRs) agonists.Eur J Med Chem. 2013 Jun;64:512-28. doi: 10.1016/j.ejmech.2013.03.066. Epub 2013 Apr 8. Eur J Med Chem. 2013. PMID: 23685570 Free PMC article.
-
4-Aroyl-3-hydroxy-5-phenyl-1H-pyrrol-2(5H)-ones as N-formyl peptide receptor 1 (FPR1) antagonists.Biochem Pharmacol. 2017 Oct 15;142:120-132. doi: 10.1016/j.bcp.2017.07.004. Epub 2017 Jul 8. Biochem Pharmacol. 2017. PMID: 28690139 Free PMC article.
-
Antagonism of human formyl peptide receptor 1 (FPR1) by chromones and related isoflavones.Biochem Pharmacol. 2014 Dec 15;92(4):627-41. doi: 10.1016/j.bcp.2014.09.027. Epub 2014 Oct 17. Biochem Pharmacol. 2014. PMID: 25450672 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

