Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis
- PMID: 22128124
- PMCID: PMC3246320
- DOI: 10.1105/tpc.111.091033
Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis
Abstract
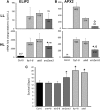
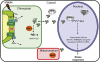
Compartmentation of the eukaryotic cell requires a complex set of subcellular messages, including multiple retrograde signals from the chloroplast and mitochondria to the nucleus, to regulate gene expression. Here, we propose that one such signal is a phosphonucleotide (3'-phosphoadenosine 5'-phosphate [PAP]), which accumulates in Arabidopsis thaliana in response to drought and high light (HL) stress and that the enzyme SAL1 regulates its levels by dephosphorylating PAP to AMP. SAL1 accumulates in chloroplasts and mitochondria but not in the cytosol. sal1 mutants accumulate 20-fold more PAP without a marked change in inositol phosphate levels, demonstrating that PAP is a primary in vivo substrate. Significantly, transgenic targeting of SAL1 to either the nucleus or chloroplast of sal1 mutants lowers the total PAP levels and expression of the HL-inducible ASCORBATE PEROXIDASE2 gene. This indicates that PAP must be able to move between cellular compartments. The mode of action for PAP could be inhibition of 5' to 3' exoribonucleases (XRNs), as SAL1 and the nuclear XRNs modulate the expression of a similar subset of HL and drought-inducible genes, sal1 mutants accumulate XRN substrates, and PAP can inhibit yeast (Saccharomyces cerevisiae) XRNs. We propose a SAL1-PAP retrograde pathway that can alter nuclear gene expression during HL and drought stress.
Figures



Comment in
-
Retrograde signaling: a new candidate signaling molecule.Plant Cell. 2011 Nov;23(11):3870. doi: 10.1105/tpc.111.231113. Epub 2011 Nov 29. Plant Cell. 2011. PMID: 22128121 Free PMC article. No abstract available.
References
-
- Allison D.B., Cui X., Page G.P., Sabripour M. (2006). Microarray data analysis: From disarray to consolidation and consensus. Nat. Rev. Genet. 7: 55–65 - PubMed
-
- Aronsson H., Jarvis P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529: 215–220 - PubMed
-
- Baldi P., Long A.D. (2001). A Bayesian framework for the analysis of microarray expression data: Regularized t -test and statistical inferences of gene changes. Bioinformatics 17: 509–519 - PubMed
-
- Ball L., Accotto G.-P., Bechtold U., Creissen G., Funck D., Jimenez A., Kular B., Leyland N., Mejia-Carranza J., Reynolds H., Karpinski S., Mullineaux P.M. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16: 2448–2462 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

