Genome-wide detection of natural selection in African Americans pre- and post-admixture
- PMID: 22128132
- PMCID: PMC3290787
- DOI: 10.1101/gr.124784.111
Genome-wide detection of natural selection in African Americans pre- and post-admixture
Abstract
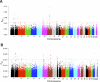
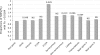
It is particularly meaningful to investigate natural selection in African Americans (AfA) due to the high mortality their African ancestry has experienced in history. In this study, we examined 491,526 autosomal single nucleotide polymorphisms (SNPs) genotyped in 5210 individuals and conducted a genome-wide search for selection signals in 1890 AfA. Several genomic regions showing an excess of African or European ancestry, which were considered the footprints of selection since population admixture, were detected based on a commonly used approach. However, we also developed a new strategy to detect natural selection both pre- and post-admixture by reconstructing an ancestral African population (AAF) from inferred African components of ancestry in AfA and comparing it with indigenous African populations (IAF). Interestingly, many selection-candidate genes identified by the new approach were associated with AfA-specific high-risk diseases such as prostate cancer and hypertension, suggesting an important role these disease-related genes might have played in adapting to a new environment. CD36 and HBB, whose mutations confer a degree of protection against malaria, were also located in the highly differentiated regions between AAF and IAF. Further analysis showed that the frequencies of alleles protecting against malaria in AAF were lower than those in IAF, which is consistent with the relaxed selection pressure of malaria in the New World. There is no overlap between the top candidate genes detected by the two approaches, indicating the different environmental pressures AfA experienced pre- and post-population admixture. We suggest that the new approach is reasonably powerful and can also be applied to other admixed populations such as Latinos and Uyghurs.
Figures
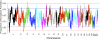
Similar articles
-
Ancestral components of admixed genomes in a Mexican cohort.PLoS Genet. 2011 Dec;7(12):e1002410. doi: 10.1371/journal.pgen.1002410. Epub 2011 Dec 15. PLoS Genet. 2011. PMID: 22194699 Free PMC article.
-
Genome-wide scan of 29,141 African Americans finds no evidence of directional selection since admixture.Am J Hum Genet. 2014 Oct 2;95(4):437-44. doi: 10.1016/j.ajhg.2014.08.011. Epub 2014 Sep 18. Am J Hum Genet. 2014. PMID: 25242497 Free PMC article.
-
Increased genetic diversity of ADME genes in African Americans compared with their putative ancestral source populations and implications for pharmacogenomics.BMC Genet. 2014 May 1;15:52. doi: 10.1186/1471-2156-15-52. BMC Genet. 2014. PMID: 24884825 Free PMC article.
-
Genetic admixture: a tool to identify diabetic nephropathy genes in African Americans.Ethn Dis. 2008 Summer;18(3):384-8. Ethn Dis. 2008. PMID: 18785456 Review.
-
Genome-wide association studies in Africans and African Americans: expanding the framework of the genomics of human traits and disease.Public Health Genomics. 2015;18(1):40-51. doi: 10.1159/000367962. Epub 2014 Nov 26. Public Health Genomics. 2015. PMID: 25427668 Free PMC article. Review.
Cited by
-
Strong selection during the last millennium for African ancestry in the admixed population of Madagascar.Nat Commun. 2018 Mar 2;9(1):932. doi: 10.1038/s41467-018-03342-5. Nat Commun. 2018. PMID: 29500350 Free PMC article.
-
Rapid adaptation to malaria facilitated by admixture in the human population of Cabo Verde.Elife. 2021 Jan 4;10:e63177. doi: 10.7554/eLife.63177. Elife. 2021. PMID: 33393457 Free PMC article.
-
Detecting Heterogeneity in Population Structure Across the Genome in Admixed Populations.Genetics. 2016 Sep;204(1):43-56. doi: 10.1534/genetics.115.184184. Epub 2016 Jul 20. Genetics. 2016. PMID: 27440868 Free PMC article.
-
Distribution of ancestral chromosomal segments in admixed genomes and its implications for inferring population history and admixture mapping.Eur J Hum Genet. 2014 Jul;22(7):930-7. doi: 10.1038/ejhg.2013.265. Epub 2013 Nov 20. Eur J Hum Genet. 2014. PMID: 24253859 Free PMC article.
-
A Comprehensive Characterization of Monoallelic Expression During Hematopoiesis and Leukemogenesis via Single-Cell RNA-Sequencing.Front Cell Dev Biol. 2021 Oct 13;9:702897. doi: 10.3389/fcell.2021.702897. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722498 Free PMC article.
References
-
- Adams J, Ward RH 1973. Admixture studies and the detection of selection. Science 180: 1137–1143 - PubMed
-
- Aitman TJ, Cooper LD, Norsworthy PJ, Wahid FN, Gray JK, Curtis BR, McKeigue PM, Kwiatkowski D, Greenwood BM, Snow RW, et al. 2000. Malaria susceptibility and CD36 mutation. Nature 405: 1015–1016 - PubMed
-
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, et al. 2009. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat Genet 41: 1058–1060 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
