Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate
- PMID: 22128180
- PMCID: PMC3268416
- DOI: 10.1074/jbc.M111.318766
Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate
Abstract
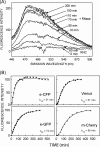
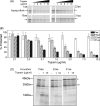
Protein folding in cells reflects a delicate interplay between biophysical properties of the nascent polypeptide, the vectorial nature and rate of translation, molecular crowding, and cellular biosynthetic machinery. To better understand how this complex environment affects de novo folding pathways as they occur in the cell, we expressed β-barrel fluorescent proteins derived from GFP and RFP in an in vitro system that allows direct analysis of cotranslational folding intermediates. Quantitative analysis of ribosome-bound eCFP and mCherry fusion proteins revealed that productive folding exhibits a sharp threshold as the length of polypeptide from the C terminus to the ribosome peptidyltransferase center is increased. Fluorescence spectroscopy, urea denaturation, and limited protease digestion confirmed that sequestration of only 10-15 C-terminal residues within the ribosome exit tunnel effectively prevents stable barrel formation, whereas folding occurs unimpeded when the C terminus is extended beyond the ribosome exit site. Nascent FPs with 10 of the 11 β-strands outside the ribosome exit tunnel acquire a non-native conformation that is remarkably stable in diverse environments. Upon ribosome release, these structural intermediates fold efficiently with kinetics that are unaffected by the cytosolic crowding or cellular chaperones. Our results indicate that during synthesis, fluorescent protein folding is initiated cotranslationally via rapid formation of a highly stable, on-pathway structural intermediate and that the rate-limiting step of folding involves autonomous incorporation of the 11th β-strand into the mature barrel structure.
Figures






Similar articles
-
Cotranslational Folding of Proteins on the Ribosome.Biomolecules. 2020 Jan 7;10(1):97. doi: 10.3390/biom10010097. Biomolecules. 2020. PMID: 31936054 Free PMC article. Review.
-
Cotranslational folding increases GFP folding yield.Biophys J. 2010 Apr 7;98(7):1312-20. doi: 10.1016/j.bpj.2009.12.4291. Biophys J. 2010. PMID: 20371331 Free PMC article.
-
Cotranslational Protein Folding inside the Ribosome Exit Tunnel.Cell Rep. 2015 Sep 8;12(10):1533-40. doi: 10.1016/j.celrep.2015.07.065. Epub 2015 Aug 28. Cell Rep. 2015. PMID: 26321634 Free PMC article.
-
Investigating the Effect of Chain Connectivity on the Folding of a Beta-Sheet Protein On and Off the Ribosome.J Mol Biol. 2018 Dec 7;430(24):5207-5216. doi: 10.1016/j.jmb.2018.10.011. Epub 2018 Oct 23. J Mol Biol. 2018. PMID: 30365950 Free PMC article.
-
How the ribosome shapes cotranslational protein folding.Curr Opin Struct Biol. 2024 Feb;84:102740. doi: 10.1016/j.sbi.2023.102740. Epub 2023 Dec 9. Curr Opin Struct Biol. 2024. PMID: 38071940 Free PMC article. Review.
Cited by
-
Beta-barrel scaffold of fluorescent proteins: folding, stability and role in chromophore formation.Int Rev Cell Mol Biol. 2013;302:221-78. doi: 10.1016/B978-0-12-407699-0.00004-2. Int Rev Cell Mol Biol. 2013. PMID: 23351712 Free PMC article. Review.
-
Local genic base composition impacts protein production and cellular fitness.PeerJ. 2018 Jan 16;6:e4286. doi: 10.7717/peerj.4286. eCollection 2018. PeerJ. 2018. PMID: 29362699 Free PMC article.
-
Genetic Code Optimization for Cotranslational Protein Folding: Codon Directional Asymmetry Correlates with Antiparallel Betasheets, tRNA Synthetase Classes.Comput Struct Biotechnol J. 2017 Aug 12;15:412-424. doi: 10.1016/j.csbj.2017.08.001. eCollection 2017. Comput Struct Biotechnol J. 2017. PMID: 28924459 Free PMC article.
-
The ribosome lowers the entropic penalty of protein folding.Nature. 2024 Sep;633(8028):232-239. doi: 10.1038/s41586-024-07784-4. Epub 2024 Aug 7. Nature. 2024. PMID: 39112704 Free PMC article.
-
Structural basis of fluorescence quenching in caspase activatable-GFP.Protein Sci. 2013 Mar;22(3):247-57. doi: 10.1002/pro.2188. Epub 2013 Jan 10. Protein Sci. 2013. PMID: 23139158 Free PMC article.
References
-
- Clark P. L. (2004) Protein folding in the cell. Reshaping the folding funnel. Trends Biochem. Sci. 29, 527–534 - PubMed
-
- Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 - PubMed
-
- Kramer G., Boehringer D., Ban N., Bukau B. (2009) The ribosome as a platform for cotranslational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 16, 589–597 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

