Biochemical and structural studies of uncharacterized protein PA0743 from Pseudomonas aeruginosa revealed NAD+-dependent L-serine dehydrogenase
- PMID: 22128181
- PMCID: PMC3265868
- DOI: 10.1074/jbc.M111.294561
Biochemical and structural studies of uncharacterized protein PA0743 from Pseudomonas aeruginosa revealed NAD+-dependent L-serine dehydrogenase
Abstract
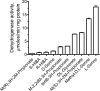
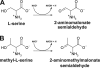
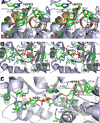
The β-hydroxyacid dehydrogenases form a large family of ubiquitous enzymes that catalyze oxidation of various β-hydroxy acid substrates to corresponding semialdehydes. Several known enzymes include β-hydroxyisobutyrate dehydrogenase, 6-phosphogluconate dehydrogenase, 2-(hydroxymethyl)glutarate dehydrogenase, and phenylserine dehydrogenase, but the vast majority of β-hydroxyacid dehydrogenases remain uncharacterized. Here, we demonstrate that the predicted β-hydroxyisobutyrate dehydrogenase PA0743 from Pseudomonas aeruginosa catalyzes an NAD(+)-dependent oxidation of l-serine and methyl-l-serine but exhibits low activity against β-hydroxyisobutyrate. Two crystal structures of PA0743 were solved at 2.2-2.3-Å resolution and revealed an N-terminal Rossmann fold domain connected by a long α-helix to the C-terminal all-α domain. The PA0743 apostructure showed the presence of additional density modeled as HEPES bound in the interdomain cleft close to the predicted catalytic Lys-171, revealing the molecular details of the PA0743 substrate-binding site. The structure of the PA0743-NAD(+) complex demonstrated that the opposite side of the enzyme active site accommodates the cofactor, which is also bound near Lys-171. Site-directed mutagenesis of PA0743 emphasized the critical role of four amino acid residues in catalysis including the primary catalytic residue Lys-171. Our results provide further insight into the molecular mechanisms of substrate selectivity and activity of β-hydroxyacid dehydrogenases.
Figures

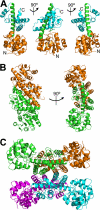

Similar articles
-
Crystal structure of novel NADP-dependent 3-hydroxyisobutyrate dehydrogenase from Thermus thermophilus HB8.J Mol Biol. 2005 Sep 30;352(4):905-17. doi: 10.1016/j.jmb.2005.07.068. J Mol Biol. 2005. PMID: 16126223
-
Crystal structure of the γ-hydroxymuconic semialdehyde dehydrogenase from Pseudomonas sp. strainWBC-3, a key enzyme involved in para-Nitrophenol degradation.BMC Struct Biol. 2013 Nov 19;13:30. doi: 10.1186/1472-6807-13-30. BMC Struct Biol. 2013. PMID: 24252642 Free PMC article.
-
Structural Insights into l-Tryptophan Dehydrogenase from a Photoautotrophic Cyanobacterium, Nostoc punctiforme.Appl Environ Microbiol. 2016 Dec 30;83(2):e02710-16. doi: 10.1128/AEM.02710-16. Print 2017 Jan 15. Appl Environ Microbiol. 2016. PMID: 27815281 Free PMC article.
-
Amine oxidation by d-arginine dehydrogenase in Pseudomonas aeruginosa.Arch Biochem Biophys. 2017 Oct 15;632:192-201. doi: 10.1016/j.abb.2017.06.013. Epub 2017 Jun 15. Arch Biochem Biophys. 2017. PMID: 28625766 Review.
-
Medium- and short-chain dehydrogenase/reductase gene and protein families : the SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes.Cell Mol Life Sci. 2008 Dec;65(24):3895-906. doi: 10.1007/s00018-008-8588-y. Cell Mol Life Sci. 2008. PMID: 19011750 Free PMC article. Review.
Cited by
-
Identification of 4-Deoxy-L-Etychro-Hexoseulose Uronic Acid Reductases in an Alginolytic Bacterium Vibrio splendidus and their Uses for L-Lactate Production in an Escherichia coli Cell-Free System.Mar Biotechnol (NY). 2018 Jun;20(3):410-423. doi: 10.1007/s10126-018-9805-9. Epub 2018 Mar 13. Mar Biotechnol (NY). 2018. PMID: 29532336
-
Crystallization and preliminary X-ray analysis of L-serine 3-dehydrogenase complexed with NADP+ from the hyperthermophilic archaeon Pyrobaculum calidifontis.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Feb 1;69(Pt 2):134-6. doi: 10.1107/S1744309112051391. Epub 2013 Jan 31. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23385753 Free PMC article.
-
Characterization of Two Late-Stage Enzymes Involved in Fosfomycin Biosynthesis in Pseudomonads.ACS Chem Biol. 2017 Feb 17;12(2):456-463. doi: 10.1021/acschembio.6b00939. Epub 2016 Dec 27. ACS Chem Biol. 2017. PMID: 27977135 Free PMC article.
-
Cloning, expression and characterization of 3-hydroxyisobutyrate dehydrogenase from Pseudomonas denitrificans ATCC 13867.PLoS One. 2013 May 1;8(5):e62666. doi: 10.1371/journal.pone.0062666. Print 2013. PLoS One. 2013. PMID: 23658760 Free PMC article.
-
Gene Expression of Putative Pathogenicity-Related Genes in Verticillium dahliae in Response to Elicitation with Potato Extracts and during Infection Using Quantitative Real-Time PCR.Pathogens. 2021 Apr 23;10(5):510. doi: 10.3390/pathogens10050510. Pathogens. 2021. PMID: 33922492 Free PMC article.
References
-
- Hawes J. W., Harper E. T., Crabb D. W., Harris R. A. (1996) Structural and mechanistic similarities of 6-phosphogluconate and 3-hydroxyisobutyrate dehydrogenases reveal a new enzyme family, the 3-hydroxyacid dehydrogenases. FEBS Lett. 389, 263–267 - PubMed
-
- Reitz S., Alhapel A., Essen L. O., Pierik A. J. (2008) Structural and kinetic properties of a beta-hydroxyacid dehydrogenase involved in nicotinate fermentation. J. Mol. Biol. 382, 802–811 - PubMed
-
- Njau R. K., Herndon C. A., Hawes J. W. (2001) New developments in our understanding of the beta-hydroxyacid dehydrogenases. Chem. Biol. Interact. 130–132, 785–791 - PubMed
-
- Lokanath N. K., Ohshima N., Takio K., Shiromizu I., Kuroishi C., Okazaki N., Kuramitsu S., Yokoyama S., Miyano M., Kunishima N. (2005) Crystal structure of novel NADP-dependent 3-hydroxyisobutyrate dehydrogenase from Thermus thermophilus HB8. J. Mol. Biol. 352, 905–917 - PubMed
-
- Adams M. J., Ellis G. H., Gover S., Naylor C. E., Phillips C. (1994) Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism. Structure 2, 651–668 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

