Dual beneficial effect of interloop disulfide bond for single domain antibody fragments
- PMID: 22128183
- PMCID: PMC3283254
- DOI: 10.1074/jbc.M111.242818
Dual beneficial effect of interloop disulfide bond for single domain antibody fragments
Abstract
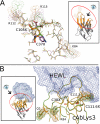
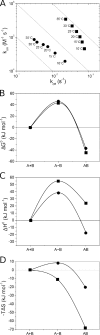
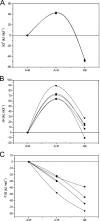
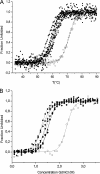
The antigen-binding fragment of functional heavy chain antibodies (HCAbs) in camelids comprises a single domain, named the variable domain of heavy chain of HCAbs (VHH). The VHH harbors remarkable amino acid substitutions in the framework region-2 to generate an antigen-binding domain that functions in the absence of a light chain partner. The substitutions provide a more hydrophilic, hence more soluble, character to the VHH but decrease the intrinsic stability of the domain. Here we investigate the functional role of an additional hallmark of dromedary VHHs, i.e. the extra disulfide bond between the first and third antigen-binding loops. After substituting the cysteines forming this interloop cystine by all 20 amino acids, we selected and characterized several VHHs that retain antigen binding capacity. Although VHH domains can function in the absence of an interloop disulfide bond, we demonstrate that its presence constitutes a net advantage. First, the disulfide bond stabilizes the domain and counteracts the destabilization by the framework region-2 hallmark amino acids. Second, the disulfide bond rigidifies the long third antigen-binding loop, leading to a stronger antigen interaction. This dual beneficial effect explains the in vivo antibody maturation process favoring VHH domains with an interloop disulfide bond.
Figures


References
-
- Williams A. F., Barclay A. N. (1988) The immunoglobulin superfamily—domains for cell surface recognition. Annu. Rev. Immunol. 6, 381–405 - PubMed
-
- Lesk A. M., Chothia C. (1982) Evolution of proteins formed by β-sheets. II. The core of the immunoglobulin domains. J. Mol. Biol. 160, 325–342 - PubMed
-
- Lefranc M. P. (2004) IMGT-ONTOLOGY and IMGT databases, tools and web resources for immunogenetics and immunoinformatics. Mol. Immunol. 40, 647–660 - PubMed
-
- Abkevich V. I., Shakhnovich E. I. (2000) What can disulfide bonds tell us about protein energetics, function and folding: simulations and bioinformatics analysis. J. Mol. Biol. 300, 975–985 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

