RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution
- PMID: 22128343
- PMCID: PMC3261745
- DOI: 10.1261/rna.02809212
RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution
Abstract
Bacterial cold shock proteins (CSPs) regulate the cellular response to temperature downshift. Their general principle of function involves RNA chaperoning and transcriptional antitermination. Here we present two crystal structures of cold shock protein B from Bacillus subtilis (Bs-CspB) in complex with either a hexanucleotide (5'-UUUUUU-3') or heptanucleotide (5'-GUCUUUA-3') single-stranded RNA (ssRNA). Hydrogen bonds and stacking interactions between RNA bases and aromatic sidechains characterize individual binding subsites. Additional binding subsites which are not occupied by the ligand in the crystal structure were revealed by NMR spectroscopy in solution on Bs-CspB·RNA complexes. Binding studies demonstrate that Bs-CspB associates with ssDNA as well as ssRNA with moderate sequence specificity. Varying affinities of oligonucleotides are reflected mainly in changes of the dissociation rates. The generally lower binding affinity of ssRNA compared to its ssDNA analog is attributed solely to the substitution of thymine by uracil bases in RNA.
Figures

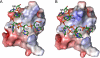
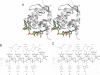
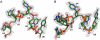

Similar articles
-
T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB.J Mol Biol. 2006 Jul 14;360(3):702-14. doi: 10.1016/j.jmb.2006.05.044. Epub 2006 Jun 2. J Mol Biol. 2006. PMID: 16780871
-
Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus.FEBS J. 2007 Mar;274(5):1265-79. doi: 10.1111/j.1742-4658.2007.05672.x. Epub 2007 Jan 31. FEBS J. 2007. PMID: 17266726
-
Recognition of T-rich single-stranded DNA by the cold shock protein Bs-CspB in solution.Nucleic Acids Res. 2006;34(16):4561-71. doi: 10.1093/nar/gkl376. Epub 2006 Sep 6. Nucleic Acids Res. 2006. PMID: 16956971 Free PMC article.
-
The cold-shock response--a hot topic.Mol Microbiol. 1994 Mar;11(5):811-8. doi: 10.1111/j.1365-2958.1994.tb00359.x. Mol Microbiol. 1994. PMID: 8022259 Review.
-
Structure and function of bacterial cold shock proteins.Cell Mol Life Sci. 2007 Jun;64(12):1457-70. doi: 10.1007/s00018-007-6388-4. Cell Mol Life Sci. 2007. PMID: 17437059 Free PMC article. Review.
Cited by
-
Purification and Characterization of a Novel Cold Shock Protein-Like Bacteriocin Synthesized by Bacillus thuringiensis.Sci Rep. 2016 Oct 20;6:35560. doi: 10.1038/srep35560. Sci Rep. 2016. PMID: 27762322 Free PMC article.
-
Characterization of Two Dinoflagellate Cold Shock Domain Proteins.mSphere. 2016 Jan 13;1(1):e00034-15. doi: 10.1128/mSphere.00034-15. eCollection 2016 Jan-Feb. mSphere. 2016. PMID: 27303711 Free PMC article.
-
Structure, stability and specificity of the binding of ssDNA and ssRNA with proteins.PLoS Comput Biol. 2019 Apr 1;15(4):e1006768. doi: 10.1371/journal.pcbi.1006768. eCollection 2019 Apr. PLoS Comput Biol. 2019. PMID: 30933978 Free PMC article.
-
Structural and catalytic insights into HoLaMa, a derivative of Klenow DNA polymerase lacking the proofreading domain.PLoS One. 2019 Apr 10;14(4):e0215411. doi: 10.1371/journal.pone.0215411. eCollection 2019. PLoS One. 2019. PMID: 30970012 Free PMC article.
-
The regulon of the RNA chaperone CspA and its auto-regulation in Staphylococcus aureus.Nucleic Acids Res. 2018 Feb 16;46(3):1345-1361. doi: 10.1093/nar/gkx1284. Nucleic Acids Res. 2018. PMID: 29309682 Free PMC article.
References
-
- Brandi A, Pon CL, Gualerzi CO 1994. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie 76: 1090–1098 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
