Integrative analyses of speciation and divergence in Psammodromus hispanicus (Squamata: Lacertidae)
- PMID: 22129245
- PMCID: PMC3293786
- DOI: 10.1186/1471-2148-11-347
Integrative analyses of speciation and divergence in Psammodromus hispanicus (Squamata: Lacertidae)
Abstract
Background: Genetic, phenotypic and ecological divergence within a lineage is the result of past and ongoing evolutionary processes, which lead ultimately to diversification and speciation. Integrative analyses allow linking diversification to geological, climatic, and ecological events, and thus disentangling the relative importance of different evolutionary drivers in generating and maintaining current species richness.
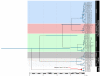
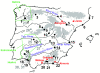
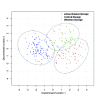
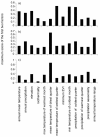
Results: Here, we use phylogenetic, phenotypic, geographic, and environmental data to investigate diversification in the Spanish sand racer (Psammodromus hispanicus). Phylogenetic, molecular clock dating, and phenotypic analyses show that P. hispanicus consists of three lineages. One lineage from Western Spain diverged 8.3 (2.9-14.7) Mya from the ancestor of Psammodromus hispanicus edwardsianus and P. hispanicus hispanicus Central lineage. The latter diverged 4.8 (1.5-8.7) Mya. Molecular clock dating, together with population genetic analyses, indicate that the three lineages experienced northward range expansions from southern Iberian refugia during Pleistocene glacial periods. Ecological niche modelling shows that suitable habitat of the Western lineage and P. h. edwardsianus overlap over vast areas, but that a barrier may hinder dispersal and genetic mixing of populations of both lineages. P. h. hispanicus Central lineage inhabits an ecological niche that overlaps marginally with the other two lineages.
Conclusions: Our results provide evidence for divergence in allopatry and niche conservatism between the Western lineage and the ancestor of P. h. edwardsianus and P. h. hispanicus Central lineage, whereas they suggest that niche divergence is involved in the origin of the latter two lineages. Both processes were temporally separated and may be responsible for the here documented genetic and phenotypic diversity of P. hispanicus. The temporal pattern is in line with those proposed for other animal lineages. It suggests that geographic isolation and vicariance played an important role in the early diversification of the group, and that lineage diversification was further amplified through ecological divergence.
Figures




References
-
- Losos JB. Integrative approaches to evolutionary ecology - Anolis lizards as model systems. Annu Rev Ecol Syst. 1994;25:467–493. doi: 10.1146/annurev.es.25.110194.002343. - DOI
-
- Wiens JJ, Graham CH. Niche conservatism: Integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst. 2005;36:519–539. doi: 10.1146/annurev.ecolsys.36.102803.095431. - DOI
-
- McCormack JE, Zellmer AJ, Knowles LL. Does niche divergence accompany allopatric divergence in Aphelocoma jays as predicted under ecological speciation?: Insights from tests with niche models. Evolution. 2010;64:1231–1244. - PubMed
-
- Pease KM, Freedman AH, Pollinger JP, McCormack JE, Buermann W, Rodzen J, Banks J, Meredith E, Bleich VC, Schaefer RJ. et al. Landscape genetics of California mule deer (Odocoileus hemionus): the roles of ecological and historical factors in generating differentiation. Mol Ecol. 2009;18(9):1848–1862. doi: 10.1111/j.1365-294X.2009.04112.x. - DOI - PubMed
-
- Mayr E. Systematics and the Origin of Species from the Viewpoint of a Zoologist. New York: Columbia University Press; 1942.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

