Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation
- PMID: 22130515
- PMCID: PMC3314179
- DOI: 10.1007/s00018-011-0884-2
Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation
Abstract
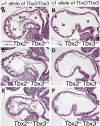
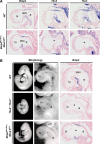
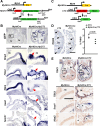
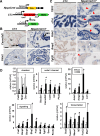
A key step in heart development is the coordinated development of the atrioventricular canal (AVC), the constriction between the atria and ventricles that electrically and physically separates the chambers, and the development of the atrioventricular valves that ensure unidirectional blood flow. Using knock-out and inducible overexpression mouse models, we provide evidence that the developmentally important T-box factors Tbx2 and Tbx3, in a functionally redundant manner, maintain the AVC myocardium phenotype during the process of chamber differentiation. Expression profiling and ChIP-sequencing analysis of Tbx3 revealed that it directly interacts with and represses chamber myocardial genes, and induces the atrioventricular pacemaker-like phenotype by activating relevant genes. Moreover, mutant mice lacking 3 or 4 functional alleles of Tbx2 and Tbx3 failed to form atrioventricular cushions, precursors of the valves and septa. Tbx2 and Tbx3 trigger development of the cushions through a regulatory feed-forward loop with Bmp2, thus providing a mechanism for the co-localization and coordination of these important processes in heart development.
Figures


References
-
- Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, Del MG, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;120:3493–3507. doi: 10.1172/JCI42666. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

