Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen and angiogenin interact with common host proteins, including annexin A2, which is essential for survival of latently infected cells
- PMID: 22130534
- PMCID: PMC3264333
- DOI: 10.1128/JVI.05754-11
Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen and angiogenin interact with common host proteins, including annexin A2, which is essential for survival of latently infected cells
Abstract
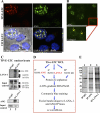
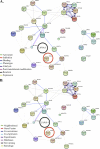
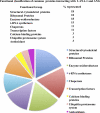
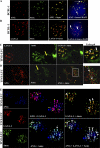
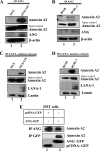
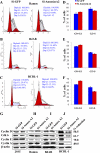
Kaposi's sarcoma-associated herpesvirus (KSHV) infection and latency-associated nuclear antigen (LANA-1) upregulate the multifunctional protein angiogenin (ANG). Our studies demonstrate that silencing ANG or inhibiting its nuclear translocation downregulates KSHV LANA-1 expression and ANG is necessary for KSHV latency, anti-apoptosis and angiogenesis (Sadagopan et al., J. Virol. 83:3342-3364, 2009; Sadagopan et al., J Virol. 85:2666-2685, 2011). Here we show that LANA-1 interacts with ANG and colocalizes in latently infected endothelial telomerase-immortalized human umbilical vein endothelial (TIVE-LTC) cells. Mass spectrometric analyses of TIVE-LTC proteins immunoprecipitated by anti-LANA-1 and ANG antibodies identified 28 common cellular proteins such as ribosomal proteins, structural proteins, tRNA synthetases, metabolic pathway enzymes, chaperons, transcription factors, antioxidants, and ubiquitin proteosome proteins. LANA-1 and ANG interaction with one of the proteins, annexin A2, was validated. Annexin A2 has been shown to play roles in cell proliferation, apoptosis, plasmin generation, exocytosis, endocytosis, and cytoskeleton reorganization. It is also known to associate with glycolytic enzyme 3-phosphoglyceratekinase in the primer recognition protein (PRP) complex that interacts with DNA polymerase α in the lagging strand of DNA during replication. A higher level of annexin A2 is expressed in KSHV+ but not in Epstein-Barr virus (EBV)+ B-lymphoma cell lines. Annexin A2 colocalized with several LANA-1 punctate spots in KSHV+ body cavity B-cell lymphoma (BCBL-1) cells. In triple-staining analyses, we observed annexin A2-ANG-LANA-1, annexin A2-ANG, and ANG-LANA-1 colocalizations. Annexin A2 appeared as punctate nuclear dots in LANA-1-positive TIVE-LTC cells. In LANA-1-negative TIVE-LTC cells, annexin A2 was detected predominately in the cytoplasm, with some nuclear spots, and colocalization with ANG was observed mostly in the cytoplasm. Annexin A2 coimmunoprecipitated with LANA-1 and ANG in TIVE-LTC and BCBL-1 cells and with ANG in 293T cells independent of LANA-1. This suggested that annexin A2 forms a complex with LANA-1 and ANG as well as a separate complex with ANG. Silencing annexin A2 in BCBL-1 cells resulted in significant cell death, downregulation of cell cycle-associated Cdk6 and of cyclin D, E, and A proteins, and downregulation of LANA-1 and ANG expression. No effect was seen in KSHV⁻ lymphoma (BJAB and Ramos) and 293T cells. These studies suggest that LANA-1 association with annexin A2/ANG could be more important than ANG association with annexin A2, and KSHV probably uses annexin A2 to maintain the viability and cell cycle regulation of latently infected cells. Since the identified LANA-1- and ANG-interacting common cellular proteins are hitherto unknown to KSHV and ANG biology, this offers a starting point for further analysis of their roles in KSHV biology, which may lead to identification of potential therapeutic targets to control KSHV latency and associated malignancies.
Figures


Similar articles
-
Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with multifunctional angiogenin to utilize its antiapoptotic functions.J Virol. 2012 Jun;86(11):5974-91. doi: 10.1128/JVI.00070-12. Epub 2012 Mar 21. J Virol. 2012. PMID: 22438557 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-induced angiogenin plays roles in latency via the phospholipase C gamma pathway: blocking angiogenin inhibits latent gene expression and induces the lytic cycle.J Virol. 2011 Mar;85(6):2666-85. doi: 10.1128/JVI.01532-10. Epub 2011 Jan 5. J Virol. 2011. PMID: 21209106 Free PMC article.
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen: Replicating and Shielding Viral DNA during Viral Persistence.J Virol. 2017 Jun 26;91(14):e01083-16. doi: 10.1128/JVI.01083-16. Print 2017 Jul 15. J Virol. 2017. PMID: 28446671 Free PMC article. Review.
-
The latency-associated nuclear antigen, a multifunctional protein central to Kaposi's sarcoma-associated herpesvirus latency.Future Microbiol. 2011 Dec;6(12):1399-413. doi: 10.2217/fmb.11.137. Future Microbiol. 2011. PMID: 22122438 Free PMC article. Review.
Cited by
-
Annexins in Translational Research: Hidden Treasures to Be Found.Int J Mol Sci. 2018 Jun 15;19(6):1781. doi: 10.3390/ijms19061781. Int J Mol Sci. 2018. PMID: 29914106 Free PMC article. Review.
-
Identification of Viral and Host Proteins That Interact with Murine Gammaherpesvirus 68 Latency-Associated Nuclear Antigen during Lytic Replication: a Role for Hsc70 in Viral Replication.J Virol. 2015 Nov 18;90(3):1397-413. doi: 10.1128/JVI.02022-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26581985 Free PMC article.
-
Cell Cycle Regulatory Functions of the KSHV Oncoprotein LANA.Front Microbiol. 2016 Mar 30;7:334. doi: 10.3389/fmicb.2016.00334. eCollection 2016. Front Microbiol. 2016. PMID: 27065950 Free PMC article. Review.
-
Nucleolin Regulates the Expression of Kaposi's Sarcoma-Associated Herpesvirus' Latency-Associated Nuclear Antigen through G-Quadruplexes in the mRNA.Viruses. 2023 Dec 15;15(12):2438. doi: 10.3390/v15122438. Viruses. 2023. PMID: 38140679 Free PMC article.
-
Molecular biology of human herpesvirus 8: novel functions and virus-host interactions implicated in viral pathogenesis and replication.Recent Results Cancer Res. 2014;193:227-68. doi: 10.1007/978-3-642-38965-8_13. Recent Results Cancer Res. 2014. PMID: 24008302 Free PMC article. Review.
References
-
- Akula SM, Pramod NP, Wang FZ, Chandran B. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407–419 - PubMed
-
- Asano K, Merrick WC, Hershey JW. 1997. The translation initiation factor eIF3-p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J. Biol. Chem. 272:23477–23480 - PubMed
-
- Babiychuk EB, Monastyrskaya K, Burkhard FC, Wray S, Draeger A. 2002. Modulating signaling events in smooth muscle: cleavage of annexin 2 abolishes its binding to lipid rafts. FASEB J. 16:1177–1184 - PubMed
-
- Bader GD, Hogue CW. 2002. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 20:991–997 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

