Murine gammaherpesvirus 68 evades host cytokine production via replication transactivator-induced RelA degradation
- PMID: 22130545
- PMCID: PMC3302405
- DOI: 10.1128/JVI.06127-11
Murine gammaherpesvirus 68 evades host cytokine production via replication transactivator-induced RelA degradation
Abstract
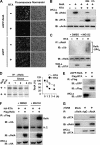
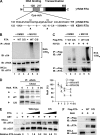
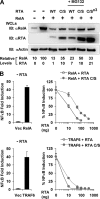
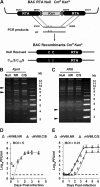
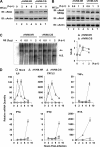
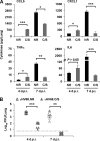
Cytokines play crucial roles in curtailing the propagation and spread of pathogens within the host. As obligate pathogens, gammaherpesviruses have evolved a plethora of mechanisms to evade host immune responses. We have previously shown that murine gammaherpesvirus 68 (γHV68) induces the degradation of RelA, an essential subunit of the transcriptionally active NF-κB dimer, to evade cytokine production. Here, we report that the immediately early gene product of γHV68, replication transactivator (RTA), functions as a ubiquitin E3 ligase to promote RelA degradation and abrogate cytokine production. A targeted genomic screen identified that RTA, out of 24 candidates, induces RelA degradation and abolishes NF-κB activation. Biochemical analyses indicated that RTA interacts with RelA and promotes RelA ubiquitination, thereby facilitating RelA degradation. Mutations within a conserved cysteine/histidine-rich, putative E3 ligase domain impaired the ability of RTA to induce RelA ubiquitination and degradation. Moreover, infection by recombinant γHV68 carrying mutations that diminish the E3 ligase activity of RTA resulted in more robust NF-κB activation and cytokine induction than did infection by wild-type γHV68. These findings support the conclusion that γHV68 subverts early NF-κB activation and cytokine production through RTA-induced RelA degradation, uncovering a key function of RTA that antagonizes the intrinsic cytokine production during gammaherpesvirus infection.
Figures

References
-
- Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801 - PubMed
-
- Ben-Neriah Y, Karin M. 2011. Inflammation meets cancer, with NF-kappaB as the matchmaker. Nat. Immunol. 12:715–723 - PubMed
-
- Borst EM, Crnkovic-Mertens I, Messerle M. 2004. Cloning of beta-herpesvirus genomes as bacterial artificial chromosomes. Methods Mol. Biol. 256:221–239 - PubMed
-
- Carbone A, Cesarman E, Spina M, Gloghini A, Schulz TF. 2009. HIV-associated lymphomas and gamma-herpesviruses. Blood 113:1213–1224 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

