Two microRNAs encoded within the bovine herpesvirus 1 latency-related gene promote cell survival by interacting with RIG-I and stimulating NF-κB-dependent transcription and beta interferon signaling pathways
- PMID: 22130548
- PMCID: PMC3264334
- DOI: 10.1128/JVI.06550-11
Two microRNAs encoded within the bovine herpesvirus 1 latency-related gene promote cell survival by interacting with RIG-I and stimulating NF-κB-dependent transcription and beta interferon signaling pathways
Abstract
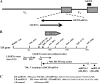
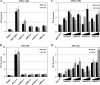
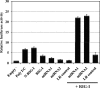
Sensory neurons latently infected with bovine herpesvirus 1 (BHV-1) abundantly express latency-related (LR) RNA (LR-RNA). Genetic evidence indicates that LR protein expression plays a role in the latency-reactivation cycle, because an LR mutant virus that contains three stop codons downstream of the first open reading frame (ORF2) does not reactivate from latency. The LR mutant virus induces higher levels of apoptotic neurons in trigeminal ganglia, and ORF2 interferes with apoptosis. Although ORF2 is important for the latency-reactivation cycle, other factors encoded by the LR gene are believed to play a supportive role. For example, two microRNAs (miRNAs) encoded within the LR gene are expressed in trigeminal ganglia of latently infected calves. These miRNAs interfere with bICP0 protein expression and productive infection in transient-transfection assays. In this report, we provide evidence that the two LR miRNAs cooperate with poly(I·C), interferon (IFN) regulatory factor 3 (IRF3), or IRF7 to stimulate beta interferon (IFN-β) promoter activity. Both miRNAs also stimulated IFN-β promoter activity and nuclear factor-kappa B (NF-κB)-dependent transcription when cotransfected with a plasmid expressing retinoic acid-inducible gene I (RIG-I). In the presence of RIG-I, the LR miRNAs enhanced survival of mouse neuroblastoma cells, which correlated with activation of the antiapoptosis cellular transcription factor, NF-κB. Immunoprecipitation assays demonstrated that both miRNAs stably interact with RIG-I, suggesting that this interaction directly stimulates the RIG-I signaling pathway. In summary, the results of these studies suggest that interactions between LR miRNAs and RIG-I promote the establishment and maintenance of latency by enhancing survival of infected neurons.
Figures


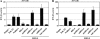


References
-
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738 - PubMed
-
- Bauman Y, et al. 2011. An identical miRNA of the human JC and BK polyoma viruses targets the stress-induced ligand ULBP3 to escape immune elimination. Cell Host Microbe 9: 93–102 - PubMed
-
- Belardo G, Santoro MG. 2010. Heat stress triggers apoptosis by impairing NF-kappaB sirvival signaling in malignant B cells. Leukemia 24: 187–196 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

