Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa
- PMID: 22130969
- PMCID: PMC3341826
- DOI: 10.1093/molbev/msr293
Evolution of functionally diverse alleles associated with PTC bitter taste sensitivity in Africa
Abstract
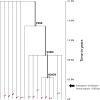
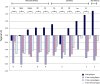
Although human bitter taste perception is hypothesized to be a dietary adaptation, little is known about genetic signatures of selection and patterns of bitter taste perception variability in ethnically diverse populations with different diets, particularly from Africa. To better understand the genetic basis and evolutionary history of bitter taste sensitivity, we sequenced a 2,975 bp region encompassing TAS2R38, a bitter taste receptor gene, in 611 Africans from 57 populations in West Central and East Africa with diverse subsistence patterns, as well as in a comparative sample of 132 non-Africans. We also examined the association between genetic variability at this locus and threshold levels of phenylthiocarbamide (PTC) bitterness in 463 Africans from the above populations to determine how variation influences bitter taste perception. Here, we report striking patterns of variation at TAS2R38, including a significant excess of novel rare nonsynonymous polymorphisms that recently arose only in Africa, high frequencies of haplotypes in Africa associated with intermediate bitter taste sensitivity, a remarkably similar frequency of common haplotypes across genetically and culturally distinct Africans, and an ancient coalescence time of common variation in global populations. Additionally, several of the rare nonsynonymous substitutions significantly modified levels of PTC bitter taste sensitivity in diverse Africans. While ancient balancing selection likely maintained common haplotype variation across global populations, we suggest that recent selection pressures may have also resulted in the unusually high level of rare nonsynonymous variants in Africa, implying a complex model of selection at the TAS2R38 locus in African populations. Furthermore, the distribution of common haplotypes in Africa is not correlated with diet, raising the possibility that common variation may be under selection due to their role in nondietary biological processes. In addition, our data indicate that novel rare mutations contribute to the phenotypic variance of PTC sensitivity, illustrating the influence of rare variation on a common trait, as well as the relatively recent evolution of functionally diverse alleles at this locus.
Figures



References
-
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. - PubMed
-
- Blum MG, Jakobsson M. Deep divergences of human gene trees and models of human origins. Mol Biol Evol. 2011;28:889–898. - PubMed
-
- Bustamante CD, Fledel-Alon A, Williamson S, et al. (14 co-authors). 2005. Natural selection on protein-coding genes in the human genome. Nature. 43:1153–1157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

